食品メタボロームレポジトリ(食レポ)は、主要な食品に含まれる成分を、 液体クロマトグラフィー-質量分析(LC-MS)によるメタボローム解析で 分析したデータを集積・公開しているサイトです。
食レポの目的:未知成分のリファレンス
私たちを取り巻く環境中には、存在は確認できるものの、未だ同定されず活用されていない
未知の化合物がたくさん存在しています。それらを同定し利活用するためには、
どの未知成分に焦点を当てて詳細な研究をしてゆくべきか、取捨選択する必要があります。
食レポでは、未知物質がどの試料に局在しているかを調べることができ、
研究対象を絞り込む大きなヒントとなります。
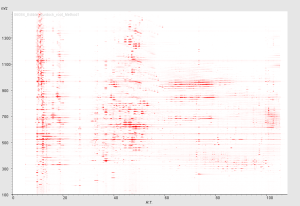
特徴1:成分の多様性を視覚化
LC-MSで検出される成分ピークのパターンを、2次元マップで視覚的に確認できます。 野菜や果物は、魚の切り身などに比べて非常に大きな成分的多様性を持っていることなどが、 よくわかります。
特徴2:精密質量値での検索
特定の質量値を持つ成分ピークが、どの食品に含まれるかを検索できます。
未知成分であっても、サンプル局在性が分かると、その成分を深く研究すべきかどうかの判断材料となります。
成分ピークのサンプル特異性が分かると、成分の推定に役立ちます。
特徴3:マススペクトルでの検索
マススペクトル(複数のイオンの質量値vs強度データ)をもとに、同様のスペクトルが検出されたピークを検索できます。
MS2(MS/MS)データに加えて、MS3データが取得されているピークもあるため、例えばフラボノイドなどの特定のマススペクトルを 示す化合物について、サンプル間での配糖体のバリエーションの有無などを確認できます。
※全てのピークについてマススペクトル情報が得られているわけではありません。
特徴4:バイオインフォマティックスでの活用
データの閲覧や検索の機能は、すべてコンピュータープログラムから直接利用することができます(API機能)。 これにより、大量のデータ検索を自動的に行い、バイオインフォマティクス(生物情報学)の手法を活用して、 高度なデータ解析が可能となります。
検索条件を含むURLにアクセスすると、結果がJSONと呼ばれるテキスト形式で得られます
詳細はヘルプの「APIによる高度な利用」をご参照ください。Pythonのサンプルコードもあります。
API: Application programming interfaceの略
JSON: JavaScript Objectの略
対象とした食品
文部科学省により公開されている 日本食品標準成分表2015年版(七訂)に含まれる約2000品目(調理方法の違いなどを含む)のうち、 代表的で入手しやすい222品目を選び、分析対象としています。
分析方法
水への溶けやすさが中程度で、分子量100~1500程度の化合物に適した分析方法です。揮発性成分や、疎水性が特に強い成分は ほとんど検出されません。メタノールで抽出し、C18カラムを用いて水-アセトニトリル系で逆相LC分離を行った後、 高分解能質量分析装置を用いて、エレクトロスプレーイオン化 (ESI)法で陽イオン分析(ポジティブモード)および陰イオン分析(ネガティブモード)を行っています。 強度の強い成分は、MS2, MS3スペクトルが得られている場合があります。
解析データ
観測された精密質量値で、化合物データベースを検索した結果を付加しています。また、 MS2, MS3スペクトルとFlavonoidSearchシステムを用いて、フラボノイドアグリコンの予測を行った結果を添付しています。
- 候補として示された成分がその食品に含まれることを保証するものではありません
- 候補にない成分が、その食品に含まれていないことを保証するものではありません
姉妹サイト
| 植物メタボロームレポジトリ | |
| 万物メタボロームレポジトリ |
 ピークをシェア
ピークをシェア
興味深いピークやスペクトル検索結果が見つかったら、検索時にブラウザに表示されるURLを
ツイートしてシェアしましょう。意外な解釈が得られるかも?
@shokurepo4 フォローよろしくお願いします。ハッシュタグ「#食レポリサーチ」または「#植レポ」