Download
MassChroBook
Sample List
A sample list with sample ID, sample name, category name, and NCBI taxonomy ID is available in the Google spreadsheet.
PowerGetBatch parameters
A set of parameter files for PowerGetBatch tool which was used for peak detection and alignment. The PowerGetBatch tool is available from the link page shown below.
PowerGetBatch parameter files. Updated: 2022/07/23
Previous data
The previous versions of the published data are available below.
We archive the repository data irregularly when we overwrite the published data.
This ensures the reproducibility of the data analysis performed with this repository.
Please note that we do not change the mass chromatogram data (.raw and .mzXML)
available in the "LC-MS data" below.
The peak file set also available on each "LC-MS data" page is irregularly updated,
and the data in the file set are not necessarily identical to the archived files.
| manual_previousData_en.pdf(301 KB) The user guide for use of the previous data. |
|
| archive_ThingMR_220305.zip(615 MB) Published: 2022/03/05, Archived: 2022/04/18 |
|
| archive_ThingMR_220418.zip(670 MB) Published: 2022/04/18, Archived: 2022/04/18 |
|
| archive_ThingMR_220625.zip(911 MB) Published: 2022/06/25, Archived: 2022/06/25 |
|
| archive_ThingMR_220723.zip(1.14 GB) Published: 2022/07/23, Archived: 2022/07/23 |
|
| archive_ThingMR_221024.zip(1.20 GB) Published: 2022/10/24, Archived: 2022/10/24 |
|
| archive_ThingMR_230217.zip(1.29 GB) Published: 2023/2/17, Archived: 2023/2/17 |
|
| archive_ThingMR_240206.zip(1.33 GB) Published: 2024/2/6, Archived: 2024/2/6 |
|
| archive_ThingMR_240822.zip(1.35 GB) Published: 2024/8/22, Archived: 2024/8/22 |
|
| archive_ThingMR_250815.zip(1.40 GB) Published: 2025/8/15, Archived: 2025/8/15 |
The aligned peak data
The term "alignment" represents here the calculation to match the same
chemical peak with a similar m/z value and retention time among the
multiple samples. The dataset provided here is the alignment results
calculated using data from all samples deposited in ThingMR as tables
containing sample vs. unique peak information.
* We release the alignment data irregularly because the calculation of
all peak data in ThingMR requires high computational cost and time.
| readme_alignmentData_en.pdf(100 KB) The user guide for use and structure of the dataset. |
|
| alignment_ThingMR_535_220415.zip(513 MB) The results using 535 samples published on March 5th, 2022. Published: 2022/07/27, Archived: 2022/04/15 |
Retention time converter
This MS Excel file provides the retention time conversion between
ThingMR and FoodMR/PlantMR.
This is for your information, too, when you convert the retention time between
XMRs and your own LC-MS system.
Note: The results are just approximated values and are not accurate.
LC-MS data
The list of detected peaks, raw mass chromatogram data (ThermoFisher .raw), and
the mass chromatogram data converted to mzXML format are available for each sample.
The mzXML files and the peak list files can be analyzed using MassChroViewer tool
which is available below.
| No. | Category | Name | Image | |
|---|---|---|---|---|
| Plant;Leaf | Japanese maple / Leaf / Acer palmatum Thunb. / Sapindaceae Acer |  |
||
| Plant;Shoot | Common henbit / Shoot / Lamium amplexicaule / Lamiaceae Lamioideae Lamium |  |
||
| Plant;Shoot | Spring star / Shoot / Ipheion uniflorum / Amaryllidaceae Allioideae Ipheion |  |
||
| Plant;Root | Spring star / Root (unwashed) / Ipheion uniflorum / Amaryllidaceae Allioideae Ipheion |  |
||
| Plant;Root | Spring star / Root (washed) / Ipheion uniflorum / Amaryllidaceae Allioideae Ipheion |  |
||
| Food;Fermented | Soysauce (ethyl acetate extract) |  |
||
| Plant;Leaf | Oleander / Leaf / Nerium oleander L. var. indium / Apocynaceae Apocynoideae Nerium |  |
||
| Plant;Leaf | Yew plum pine / Leaf / Podocarpus macrophyllus lamb. / Podocarpaceae Podocarpus |  |
||
| Plant;Leaf | Loquat / Leaf / Eriobotrya japonica / Rosaceae Maloideae Eriobotrya |  |
||
| Plant;Leaf | Japanese spindle / Leaf / Euonymus japonicus / Celastraceae Euonymus |  |
||
| Plant;Fruit;Food | Tomato / Mature fruit (flesh, peel) / Solanum lycopercisum / Solanaceae Solanum |  |
||
| Plant;Leaf | Mugwort / Leaf / Artemisia indica Willd. var. maximowiczii / Asteraceae Asteroideae Artemisia |  |
||
| Plant;Root | Mugwort / Root (unwashed) / Artemisia indica Willd. var. maximowiczii / Asteraceae Asteroideae Artemisia |  |
||
| Plant;Root | Mugwort / Root (washed) / Artemisia indica Willd. var. maximowiczii / Asteraceae Asteroideae Artemisia |  |
||
| Plant;Shoot | Big quaking grass / Shoot / Briza maxima L. / Poaceae Briza |  |
||
| Plant;Root | Big quaking grass / Root (unwashed) / Briza maxima L. / Poaceae Briza |  |
||
| Plant;Shoot | Rosemary / Leaf, Stem / Rosmarinus officinalis L. / Lamiaceae Rosmarinus |  |
||
| Plant;Leaf | Dokudami / Leaf / Houttuynia cordata / Saururaceae Houttuynia |  |
||
| Plant;Shoot | Field horsetail / Shoot / Equisetum arvense / Equisetaceae Equisetum |  |
||
| Plant;Leaf | Hydrangea / Leaf / Hydrangea macrophylla / Hydrangeaceae Hydrangea |  |
||
| Plant;Shoot | Black medic / Shoot / Medicago lupulina / Fabaceae Fabiodeae Medicago |  |
||
| Plant;Shoot | Commonfield speedwell / Shoot / Veronica persica / Plantaginaceae Veronica |  |
||
| Plant;Shoot | Shepherd's purse / Shoot / Capsella bursa-pastoris / Brassicaceae Capsella |  |
||
| Plant;Shoot | Bog yellowcress / Shoot / Rorippa palustris / Brassicaceae Rorippa |  |
||
| Plant;Shoot | Rescue grass / Shoot / Bromus catharticus Vahl / Poaceae Bromus |  |
||
| Plant;Shoot | Canada toadflax / Shoot / Nuttallanthus canadensis / Plantaginaceae Antirrhineae Nuttallanthus |  |
||
| Plant;Root | Canada toadflax / Root (unwashed) / Nuttallanthus canadensis / Plantaginaceae Antirrhineae Nuttallanthus |  |
||
| Plant;Shoot | Orange foxtail / Shoot / Alopecurus aequalis / Poaceae Alopecurus |  |
||
| Plant;Root | Orange foxtail / Root (unwashed) / Alopecurus aequalis / Poaceae Alopecurus |  |
||
| Plant;Root | Orange foxtail / Root (washed) / Alopecurus aequalis / Poaceae Alopecurus |  |
||
| Plant;Shoot | Wood sorrel / Shoot / Oxalis corniculata or Oxalis dillenii / Oxalidaceae Oxalis |  |
||
| Plant;Root | Wood sorrel / Root (unwashed) / Oxalis corniculata or Oxalis dillenii / Oxalidaceae Oxalis |  |
||
| Plant;Shoot | Chinese milk vetch / Shoot / Astragalus sinicus L. / Fabaceae Faboideae Astragalus |  |
||
| Plant;Root | Chinese milk vetch / Root (unwashed) / Astragalus sinicus L. / Fabaceae Faboideae Astragalus |  |
||
| Plant;Root | Chinese milk vetch / Root (washed) / Astragalus sinicus L. / Fabaceae Faboideae Astragalus |  |
||
| Plant;Shoot | Long-headed poppy / Shoot / Papaver dubium L. / Papaveraceae Papaver |  |
||
| Plant;Root | Long-headed poppy / Root (unwashed) / Papaver dubium L. / Papaveraceae Papaver |  |
||
| Plant;Root | Long-headed poppy / Root (washed) / Papaver dubium L. / Papaveraceae Papaver |  |
||
| Plant;Leaf | Japanese Aucuba / Leaf / Aucuba japonica / Garryaceae Aucuba |  |
||
| Plant;Leaf | Canary Island ivy (putative) / Leaf / Hedera canariensis / Araliaceae Hedera |  |
||
| Plant;Shoot | Hop / Leaf, Stem / Humulus lupulus / Cannabaceae Humulus |  |
||
| Plant | Hop / Underground stem, Root, Young shoot (unwashed) / Humulus lupulus / Cannabaceae Humulus |  |
||
| Plant;Root | Hop / Underground stem, Root (unwashed) / Humulus lupulus / Cannabaceae Humulus |  |
||
| Plant;Root | Hop / Underground stem, Root (washed) / Humulus lupulus / Cannabaceae Humulus |  |
||
| Plant;Leaf | Aloe / Leaf / Aloe arborescens / Xanthorrhoeaceae Asphodeloideae Aloe |  |
||
| Plant;Flower | Linaria Ripple Stone / Flower / Linaria aeruginea 'Neon lights'/ Plantaginaceae Antirrhineae Linaria |  |
||
| Plant;Shoot | Linaria Ripple Stone / Leaf, Stem / Linaria aeruginea 'Neon lights'/ Plantaginaceae Antirrhineae Linaria |  |
||
| Plant;Flower | Marguerite Fruity pink / Flower / Argyranthemum frutescens / Asteraceae Asteroideae Argyranthemum |  |
||
| Plant;Shoot | Marguerite Fruity pink / Leaf, Stem / Argyranthemum frutescens / Asteraceae Asteroideae Argyranthemum |  |
||
| Plant;Flower | Nemophila, Baby blue eyes / Flower / Nemophila menziesii / Boraginaceae Nemophila |  |
||
| Plant;Shoot | Nemophila, Baby blue eyes / Leaf, Stem / Nemophila menziesii / Boraginaceae Nemophila |  |
||
| Plant;Flower;Food | Broccoli / Flower (edible part) / Brassica oleracea var. italica / Brassicaceae Brassica |  |
||
| Plant;Leaf;Food | Perilla (Ao-shiso) / Leaf / Perilla frutescens var. crispa / Lamiaceae Perilla |  |
||
| Herbal medicine;Plant | Kakkonto (Extract Granules) |  |
||
| Plant;Flower | Lavender (STOECHAS putative) / Flower / Lavandula sp. / Lamiaceae Lavandula |  |
||
| Plant;Shoot | Lavender (STOECHAS putative) / Leaf, Stem / Lavandula sp. / Lamiaceae Lavandula |  |
||
| Plant;Flower | Snapdragon / Flower / Antirrhinum majus L. / Plantaginaceae Antirrhinum |  |
||
| Plant;Shoot | Snapdragon / Leaf, Stem / Antirrhinum majus L. / Plantaginaceae Antirrhinum |  |
||
| Plant;Flower | Brachyscome Brasco Violet / Flower / Brachyscome angustifolia Brasco (TM) Violet / Asteraceae Brachyscome |  |
||
| Plant;Shoot | Brachyscome Brasco Violet / Leaf, Stem / Brachyscome angustifolia Brasco (TM) Violet / Asteraceae Brachyscome |  |
||
| Plant;Flower | Geranium / Flower / Pelargonium x hortorum / Geraniaceae Pelargonium |  |
||
| Plant;Shoot | Geranium / Leaf, Petiole / Pelargonium x hortorum / Geraniaceae Pelargonium |  |
||
| Plant;Flower | Yellow daisy / Flower / Chrysanthemum multicaule Desf. / Asteraceae Chrysanthemum |  |
||
| Plant;Shoot | Yellow daisy / Leaf, Stem / Chrysanthemum multicaule Desf. / Asteraceae Chrysanthemum |  |
||
| Plant;Flower | Daisy / Flower / Bellis caerulescens / Asteraceae Bellis |  |
||
| Plant;Leaf | Daisy / Leaf / Bellis caerulescens / Asteraceae Bellis |  |
||
| Plant;Waterweed; Shoot | Gasha-moku / Leaf, Stem / Potamogeton dentatus Hagstr. / Potamogetonaceae Potamogeton |  |
||
| Plant;Waterweed; Shoot | Curly-leaf pondweed / Leaf, Stem / Potamogeton crispus L. / Potamogetonaceae Potamogeton |  |
||
| Plant;Waterweed;Shoot;Root | Eurasian watermilfoil / Leaf, Stem, Root / Myriophyllum spicatum / Haloragaceae Myriophyllum |  |
||
| Environment;Waterweed | Gasha-moku-cultured water |  |
||
| Environment;Waterweed | Braun's stonewort (Chara braunii) cultured water |  |
||
| Plant;Flower | Marigold / Flower / Tagetes L / Asteraceae Asteroideae Tagetes |  |
||
| Plant;Leaf | Marigold / Leaf / Tagetes L / Asteraceae Asteroideae Tagetes |  |
||
| Plant;Flower | Gazania Gazoo / Flower / Gazania rigens / Asteraceae Cichorioideae Gazania |  |
||
| Plant;Leaf | Gazania Gazoo / Leaf / Gazania rigens / Asteraceae Cichorioideae Gazania |  |
||
| Plant;Flower | Dalmatian bellflower / Flower / Campanula portenschlagiana / Campanulaceae Campanula |  |
||
| Plant;Shoot | Dalmatian bellflower / Leaf, Stem, Flower bud (young) / Campanula portenschlagiana / Campanulaceae Campanula |  |
||
| Plant;Flower | Lobelia / Flower / Lobelia L. / Campanulaceae Lobelia |  |
||
| Plant;Shoot | Lobelia / Flower / Leaf, Stem, Flower bud (small) / Campanulaceae Lobelia |  |
||
| Plant;Flower | Osteospermum Grand Canyon / Flower / Osteospermum hybrids / Asteraceae Asteroidear Calenduleae Osteospermum |  |
||
| Plant;Leaf | Osteospermum Grand Canyon / Leaf / Osteospermum hybrids / Asteraceae Asteroidear Calenduleae Osteospermum |  |
||
| Plant;Shoot | Dwarf baby's breath / Flower, Leaf, Stem / Gypsophila cerastioides / Caryophyllaceae Gypsophila |  |
||
| Plant;Shoot | Moss phlox / Flower, Leaf, Stem/ Phlox subulata / Polemoniaceae Phlox |  |
||
| Plant;Shoot | Scrambling gromwell / Flower, Leaf, Stem, Flower bud / Glandora diffusa / Boraginaceae Glandora |  |
||
| Plant;Flower | Petunia Blue / Flower / Petunia x hybrida / Solanaceae Petunia |  |
||
| Plant;Shoot | Petunia Blue / Leaf, Stem, Flower bud (small) / Petunia x hybrida / Solanaceae Petunia |  |
||
| Plant;Flower | Lavender Furano (SPICA putative)/ Flower, Flower bud / Lavandula sp. / Lamiaceae Lavandula |  |
||
| Plant;Shoot | Lavender Furano (SPICA putative)/ Leaf, Stem, Flower bud / Lavandula sp. / Lamiaceae Lavandula |  |
||
| Plant;Root | Nemophila, Baby blue eyes / Root (washed) / Nemophila menziesii / Boraginaceae Nemophila |  |
||
| Herbal medicine;Plant | Kakkontokasenkyushin'i (Extract Granules) |  |
||
| Standard Chemical | Standard mix. / Gallic acid; Caffeic acid; 3-(3,4-dihydroxyphenyl)propanoic acid (DPA); Caffeine; Cinchonine; Thiamine; L-Glutathione; Hydroquinidine; Strychnine; Norlaudanosine; Amygdalin |  |
||
| Standard Chemical | Standard mix. / Gramine; Theophylline; Sedanolide; Genistein; Cinchonidine; Quercetin; Quinine; Sinomenine; Papaverine; Brucine; Loperamide |  |
||
| Plant;Leaf | Wild Korean mulberry (putative) / Leaf / Morus australis (putative) / Moraceae Morus |  |
||
| Plant;Flower | Wild Korean mulberry (putative) / Female flower / Morus australis (putative) / Moraceae Morus |  |
||
| Plant;Flower | Azalea / Flower / Rhododendron ferrugineum L. / Ericaceae Rhododendron |  |
||
| Plant;Shoot | Azalea / Leaf, Stem / Rhododendron ferrugineum L. / Ericaceae Rhododendron |  |
||
| Plant;Flower | Cherry blossom Itokukuri / Flower / Cerasus Sato-zakura Group 'Nobilis' / Rosaceae Amygdaloideae Cerasus |  |
||
| Plant;Shoot | Carolina geranium / Leaf, Petiole / Geranium carolinianum / Geraniaceae Geranium |  |
||
| Plant;Leaf | Ring-cup aok (putative) / Leaf / Quercus glauca / Fagaceae Quercus |  |
||
| Plant;Flower | Dandelion / Flower / Taraxacum officinale / Asteraceae Taraxacum |  |
||
| Plant;Shoot | Dandelion / Leaf, Stem / Taraxacum officinale / Asteraceae Taraxacum |  |
||
| Plant;Leaf | Japanese knotweed (putative) / Leaf / Fallopia japonica / Polygonaceae Fallopia |  |
||
| Plant;Flower | Cobra lily Urashima / Flower / Arisaema urashima / Araceae Arisaema |  |
||
| Plant;Leaf | Cobra lily Urashima / Leaf / Arisaema urashima / Araceae Arisaema |  |
||
| Plant;Flower | Satsuki azalea / Flower / Rhododendron indicum / Ericaceae Rhododendron |  |
||
| Plant;Leaf | Satsuki azalea / Leaf / Rhododendron indicum / Ericaceae Rhododendron |  |
||
| Plant;Flower | Bigleaf periwinkle / Flower / Vinca major / Apocynaceae Rauvolfioideae Vinceae Vinca |  |
||
| Plant;Shoot | Bigleaf periwinkle / Leaf, Stem / Vinca major / Apocynaceae Rauvolfioideae Vinceae Vinca |  |
||
| Plant;Flower | Variety of Raspberry / Flower / Rubus hirsutus / Rosaceae Rosoideae Rubus |  |
||
| Plant;Leaf | Variety of Raspberry / Leaf / Rubus hirsutus / Rosaceae Rosoideae Rubus |  |
||
| Herbal medicine;Plant | Otsujito (Extract Granules) |  |
||
| Plant;Shoot | Petty spurge / Shoot / Euphorbia peplus / Euphorbiaceae Euphorbia |  |
||
| Plant;Shoot | Chocolate vine / Leaf, Petiole / Akebia quinata / Lardizabalaceae Akebia |  |
||
| Plant;Shoot | An Angelica species / Leaf, Petiole / Angelica sp. / Apiaceae Angelica |  |
||
| Plant;Flower | Philadelphia fleabane / Flower / Erigeron philadelphicus L. / Asteraceae Asteroideae Astereae Erigeron |  |
||
| Plant;Shoot | Philadelphia fleabane / Leaf, Stem / Erigeron philadelphicus L. / Asteraceae Asteroideae Astereae Erigeron |  |
||
| Plant;Leaf | Ginkgo / Leaf / Ginkgo biloba L. / Ginkgoaceae Ginkgo |  |
||
| Plant;Flower | Ginkgo / Male flower / Ginkgo biloba L. / Ginkgoaceae Ginkgo |  |
||
| Plant;Shoot | Dawn redwood / Leaf, Stem / Metasequoia glyptostroboides / Cupressaceae Metasequoia |  |
||
| Plant;Leaf | Japanese Mallotus / Leaf / Mallotus japonicus / Euphorbiaceae Acalyphoideae Acalypheae Mallotus |  |
||
| Plant;Root | Geranium / Root (washed) / Pelargonium x hortorum / Geraniaceae Pelargonium |  |
||
| Herbal medicine;Plant | Anchusan (Extract Granules) |  |
||
| Herbal medicine;Plant | Jumihaidokuto (Extract Granules) |  |
||
| Herbal medicine;Plant | Hachimijiogan (Extract Granules) |  |
||
| Standard Chemical | Standard mix. Amino acids / L-alpha-Alanine; L(+)-Arginine; L-Asparagine; L-Aspartic acid; L-Cysteine; L-Glutamine; L-Glutamic acid; Glycine; L-Histidine; L-(+)-Isoleucine; L-Leucine; L-Lysine; L-Methionine; L-(-)-Phenylalanine; L-(-)-Proline; L-Seri |  |
||
| Standard Chemical | Standard L-(+)-Isoleucine |  |
||
| Animal;Blood | Human / Plasma / Homo sapiens sapiens / Hominidae Homininae Hominini Hominina Homo |  |
||
| Standard Chemical | Standard Saponin (from Quillaja) |  |
||
| Standard Chemical | Standard Curcumin |  |
||
| Food | Turmeric drink |  |
||
| Plant;Root | Snapdragon / Root (washed) / Antirrhinum majus L. / Plantaginaceae Antirrhinum |  |
||
| Plant;Root | Daisy / Root (washed) / Bellis caerulescens / Asteraceae Bellis |  |
||
| Plant;Root | Lobelia / Root (washed) / Lobelia L. / Campanulaceae Lobelia |  |
||
| Plant;Root | Dalmatian bellflower / Root (washed) / Campanula portenschlagiana / Campanulaceae Campanula |  |
||
| Plant;Root | Osteospermum Grand Canyon / Root (washed) / Osteospermum hybrids / Asteraceae Asteroidear Calenduleae Osteospermum |  |
||
| Plant;Root | Moss phlox / Root (washed) / Phlox subulata / Polemoniaceae Phlox |  |
||
| Plant;Root | Marigold / Root (unwashed) / Tagetes L / Asteraceae Asteroideae Tagetes |  |
||
| Plant;Root | Marigold / Root (washed) / Tagetes L / Asteraceae Asteroideae Tagetes |  |
||
| Plant;Root | Gazania Gazoo / Root (washed) / Gazania rigens / Asteraceae Cichorioideae Gazania |  |
||
| Plant;Root | Lavender Furano (SPICA putative) / Root (washed) / Lavandula sp. / Lamiaceae Lavandula |  |
||
| Plant;Root | Scrambling gromwell / Root (washed) / Glandora diffusa / Boraginaceae Glandora |  |
||
| Plant;Root | Marguerite Fruity pink / Root (washed) / Argyranthemum frutescens / Asteraceae Asteroideae Argyranthemum |  |
||
| Plant;Root | Linaria Ripple Stone / Root (washed) / Linaria aeruginea 'Neon lights' / Plantaginaceae Antirrhineae Linaria |  |
||
| Plant;Root | Lavender (STOECHAS putative) / Root (washed) / Lavandula sp. / Lamiaceae Lavandula |  |
||
| Plant;Root | Petunia Blue / Root (washed) / Petunia x hybrida / Solanaceae Petunia |  |
||
| Plant;Root | Yellow daisy / Root (washed) / Chrysanthemum multicaule Desf. / Asteraceae Chrysanthemum |  |
||
| Plant;Root | Brachyscome Brasco Violet / Root (washed) / Brachyscome angustifolia Brasco (TM) Violet / Asteraceae Brachyscome |  |
||
| Plant;Root | Dwarf baby's breath / Root (washed) / Gypsophila cerastioides / Caryophyllaceae Gypsophila |  |
||
| Food;Plant | Hop pellet / Humulus lupulus |  |
||
| Plant;Leaf | Rice / Leaf blade / Oryza sativa L. cv. Nipponbare / Poaceae Oryzoideae Oryza |  |
||
| Plant;Root | Rice / Root (unwashed) / Oryza sativa L. cv. Nipponbare / Poaceae Oryzoideae Oryza |  |
||
| Plant;Root | Rice / Root (washed) / Oryza sativa L. cv. Nipponbare / Poaceae Oryzoideae Oryza |  |
||
| Environment | Soil near by rice (Nipponbare) root |  |
||
| Environment | Soil from paddy field |  |
||
| Environment | Water from paddy field |  |
||
| Animal | Apple snail / Pomacea canaliculata / Ampullariidae Pomacea |  |
||
| Plant;Shoot | Chinese fringe bush / Leaf, Stem / Loropetalum chinense var. rubra / Hamamelidaceae Loropetalum |  |
||
| Plant;Shoot | A kind of Japanese Box (putative) / Leaf, Stem (young) / Buxus / Buxaceae Buxus |  |
||
| Plant;Leaf | Cherry blossom Karamizakura (putative) / Leaf / Cerasus pseudocerasus (Lindley) D.Don / Rosaceae Cerasus |  |
||
| Plant;Fruit | Cherry blossom Karamizakura (putative) / Fruit (immature) / Cerasus pseudocerasus (Lindley) D.Don / Rosaceae Cerasus |  |
||
| Plant;Flower | Chinese redbud / Flower (old) / Cercis chinensis / Fabaceae Caesalpinioideae Cercis |  |
||
| Plant;Leaf | Chinese redbud / Leaf / Cercis chinensis / Fabaceae Caesalpinioideae Cercis |  |
||
| Plant;Leaf | Tabunoki (Red Nanmu) (putative) / Leaf (young, red) / Machilus thunbergii / Lauraceae Machilus |  |
||
| Plant;Leaf | Tabunoki (Red Nanmu) (putative) / Leaf / Machilus thunbergii / Lauraceae Machilus |  |
||
| Plant;Leaf | Kiwifruit / Leaf, Stem / Actinidia chinensis Planch. / Actinidiaceae Actinidia |  |
||
| Plant;Shoot | Lesser quaking grass / Shoot / Briza minor / Poaceae Briza |  |
||
| Plant;Leaf | Persimmon / Leaf (young) / Diospyros kaki Thunb. / Ebenaceae Diospyros |  |
||
| Plant;Leaf | Japanese star anse (putative) / Leaf, Stem, Bud (young), [damaged] / Illicium anisatum / Schisandraceae Illicium |  |
||
| Plant;Leaf | Chinese holly / Leaf (young) / Osmanthus heterophyllus / Oleaceae Osmanthus |  |
||
| Plant;Flower | Blue daze / Flower / Evolvulus pilosus / Convolvulaceae Evolvulus |  |
||
| Plant;Shoot | Blue daze / Leaf, Stem, Flower bud / Evolvulus pilosus / Convolvulaceae Evolvulus |  |
||
| Plant;Root | Blue daze / Root (washed) / Evolvulus pilosus / Convolvulaceae Evolvulus |  |
||
| Plant;Flower | Busy Lizzie / Flower, Flower bud / Impatiens walleriana / Balsaminaceae Impatiens |  |
||
| Plant;Shoot | Busy Lizzie / Leaf, Stem, Flower bud / Impatiens walleriana / Balsaminaceae Impatiens |  |
||
| Plant;Flower | Gold Coin daisy / Flower, Flower bud / Pallenis maritima / Asteraceae Inuleae Pallenis |  |
||
| Plant;Shoot | Gold Coin daisy / Leaf, Stem / Pallenis maritima / Asteraceae Inuleae Pallenis |  |
||
| Plant;Flower | Perpetual begonia / Flower / Begonia semperflorens / Begoniaceae Begonia |  |
||
| Plant;Leaf | Perpetual begonia / Leaf / Begonia semperflorens / Begoniaceae Begonia |  |
||
| Plant;Flower | Madagascar periwinkle / Flower, Flower bud / Catharanthus roseus / Apocynaceae Rauvolfioideae Vinceae Catharanthus |  |
||
| Plant;Shoot | Madagascar periwinkle / Leaf, Stem, Flower bud (small) / Catharanthus roseus / Apocynaceae Rauvolfioideae Vinceae Catharanthus |  |
||
| Herbal medicine;Plant | Daisaikoto (Extract Granules) |  |
||
| Herbal medicine;Plant | Shosaikoto (Extract Granules) |  |
||
| Herbal medicine;Plant | Saikokeishito (Extract Granules) |  |
||
| Herbal medicine;Plant | Saikokeishikankyoto (Extract Granules) |  |
||
| Plant;Root | Busy Lizzie / Root (washed) / Impatiens walleriana / Balsaminaceae Impatiens |  |
||
| Plant;Root | Gold Coin daisy / Root (washed) / Pallenis maritima / Asteraceae Inuleae Pallenis |  |
||
| Plant;Flower | Bacopa Scopia / Flower / Chaenostoma cordatum / Scrophulariaceae Chaenostoma |  |
||
| Plant;Shoot | Bacopa Scopia / Leaf, Stem, Flower bud (small) / Chaenostoma cordatum / Scrophulariaceae Chaenostoma |  |
||
| Plant;Root | Bacopa Scopia / Root (washed) / Chaenostoma cordatum / Scrophulariaceae Chaenostoma |  |
||
| Plant;Flower | Scarlet sage / Flower / Salvia splendens / Lamiaceae Nepetoideae Mentheae Salvia |  |
||
| Plant;Leaf | Scarlet sage / Leaf, Petiole / Salvia splendens / Lamiaceae Nepetoideae Mentheae Salvia |  |
||
| Plant;Flower | Verbena / Flower / Verbena / Verbenaceae Verbeneae Verbena |  |
||
| Plant;Shoot | Verbena / Leaf, Stem / Verbena / Verbenaceae Verbeneae Verbena |  |
||
| Plant;Flower | Dianthus Olivia / Flower, Flower bud / Dianthus / Caryophyllaceae Dianthus |  |
||
| Plant;Leaf | Dianthus Olivia / Leaf, Stem / Dianthus / Caryophyllaceae Dianthus |  |
||
| Plant;Flower | Dianthus / Flower / Dianthus / Caryophyllaceae Dianthus |  |
||
| Plant;Leaf | Dianthus / Leaf, Stem / Dianthus / Caryophyllaceae Dianthus |  |
||
| Plant;Shoot | Euphorbia Snow Blizzard / Flower, Leaf, Stem / Euphorbia L. / Euphorbiaceae Euphorbioideae Euphorbia |  |
||
| Plant;Flower | Nemesia Nesia / Flower / Nemesia / Scrophulariaceae Nemesia |  |
||
| Plant;Shoot | Nemesia Nesia / Leaf, Stem, Flower bud (small) / Nemesia / Scrophulariaceae Nemesia |  |
||
| Plant;Root | Nemesia Nesia / Root (washed) / Nemesia / Scrophulariaceae Nemesia |  |
||
| Plant;Shoot | Cockscomb / Shoot (seedling) / Celosia argentea / Amaranthaceae Celosia |  |
||
| Product | Cigarette / Dried leaf part |  |
||
| Food | Honey 1 |  |
||
| Food | Honey 2 |  |
||
| Food | Honey 3 |  |
||
| Food | Honey 4 |  |
||
| Food | Honey 5 |  |
||
| Food | Honey 6 |  |
||
| Food | Honey 7 |  |
||
| Standard Chemical | Standard Berberine |  |
||
| Plant;Shoot | Kousa Dogwood / Leaf, Flower (young) / Cornus kousa / Cornaceae Cornus |  |
||
| Plant;Shoot | Japanese spindle / Leaf (young), Flower / Euonymus japonicus / Celastraceae Euonymus |  |
||
| Plant;Shoot | Camphor Laurel / Leaf, flower bud / Cinnamomum camphora / Lauraceae Cinnamomum |  |
||
| Plant;Shoot | Japanese hawthorn / Flower, Flower stalk, Leaf, Flower bud / Ixeris stolonifera / Asteraceae Cichorioideae Ixeris |  |
||
| Plant;Shoot | Red clover / Flower, Leaf, Stem, Flower bud / Trifolium pratense / Fabaceae Trifolium |  |
||
| Plant;Shoot | White clover / Flower, Shoot / Trifolium repens / Fabaceae Trifolium |  |
||
| Plant;Shoot | Hydrangea / Leaf, Flower bud / Hydrangea macrophylla / Hydrangeaceae Hydrangea |  |
||
| Plant;Shoot | Carolina geranium / Flower, Flower bud, Leaf / Geranium carolinianum / Geraniaceae Geranium |  |
||
| Plant;Shoot | Narrow-leaved Vetch / Shoot / Vicia sativa subsp. nigra / Fabaceae Vicia |  |
||
| Plant;Shoot | Rose evening primrose / Shoot / Oenothera rosea / Onagraceae Onagroideae Onagreae Oenothera |  |
||
| Plant;Shoot | Violet wood-sorrel / Shoot / Oxalis debilis / Oxalidaceae Oxalis |  |
||
| Plant;Root | Violet wood-sorrel / Bulb (washed) / Oxalis debilis / Oxalidaceae Oxalis |  |
||
| Plant;Waterweed | Large-flowered Waterweed / Leaf, Stem / Egeria densa / Hydrocharitaceae Egeria |  |
||
| Animal | Grape clearwing moth / Larva / Nokona regalis / Sesiidae Sesiinae Nokona |  |
||
| Animal | Earthrowm (for fishing) |  |
||
| Animal | Penis fish / Urechis unicinctus / Urechidae Urechis |  |
||
| Animal | Ragworm (Aoisome) / Perinereis aibuhitensis (putative) / Nereididae Perinereis |  |
||
| Animal | Ragworm (Akaisome) / Perinereis aibuhitensis (putative) / Nereididae Perinereis |  |
||
| Animal | Ragworm (Gold) / Perinereis aibuhitensis (putative) / Nereididae Perinereis |  |
||
| Herbal medicine;Plant | Saikokaryukotsuboreito (Extract Granules) |  |
||
| Herbal medicine;Plant | Hangeshashinto (Extract Granules) |  |
||
| Herbal medicine;Plant | Orengedokuto (Extract Granules) |  |
||
| Herbal medicine;Plant | Hangekobokuto (Extract Granules) |  |
||
| Herbal medicine;Plant | Goreisan (Extract Granules) |  |
||
| Herbal medicine;Plant | Keishikajutsubuto (Extract Granules) |  |
||
| Herbal medicine;Plant | Shoseiryuto (Extract Granules) |  |
||
| Herbal medicine;Plant | Boiogito (Extract Granules) |  |
||
| Herbal medicine;Plant | Shohangekabukuryoto (Extract Granules) |  |
||
| Herbal medicine;Plant | Shofusan (Extract Granules) |  |
||
| Herbal medicine;Plant | Tokishakuyakusan (Extract Granules) |  |
||
| Herbal medicine;Plant | Kamishoyosan (Extract Granules) |  |
||
| Herbal medicine;Plant | Keishibukuryogan (Extract Granules) |  |
||
| Herbal medicine;Plant | Keishikaryukotsuboreito (Extract Granules) |  |
||
| Herbal medicine;Plant | Maoto (Extract Granules) |  |
||
| Herbal medicine;Plant | Eppikajutsuto (Extract Granules) |  |
||
| Plant;Leaf | Cherry blossom Yamazakura-shidare / Leaf / Cerasus Sato-zakura Group 'Sendai-shidare' / Rosaceae Cerasus |  |
||
| Plant;Leaf | Cherry blossom Hayazaki-oshima / Leaf / Cerasus x yedoensis (Matsumura) A. Vassiliev / Rosaceae Cerasus |  |
||
| Plant;Leaf | Cherry blossom Usubeni-kanzakura / Leaf / Cerasus x kanzakura 'Praecox' / Rosaceae Cerasus |  |
||
| Plant;Leaf | Cherry blossom Somei-yoshino / Leaf / Cerasus x yedoensis 'Somei-yoshino' / Rosaceae Cerasus |  |
||
| Plant;Leaf | Cherry blossom Yedo-higan / Leaf / Cerasus spachiana Lavalle'e ex H.Otto / Rosaceae Cerasus |  |
||
| Plant;Leaf | Cherry blossom Oshima-zakura / Leaf / Cerasus speciosa (Koidz.) H.Ohba / Rosaceae Cerasus |  |
||
| Plant;Root | Cockscomb / Root (washed) / Celosia argentea / Amaranthaceae Celosia |  |
||
| Plant;Root | Euphorbia Snow Blizzard / Root (washed) / Euphorbia L. / Euphorbiaceae Euphorbioideae Euphorbia |  |
||
| Plant;Root | Madagascar periwinkle / Root (washed) / Catharanthus roseus / Apocynaceae Rauvolfioideae Vinceae Catharanthus |  |
||
| Plant;Root | Perpetual begonia / Root (washed) / Begonia semperflorens / Begoniaceae Begonia |  |
||
| Plant;Root | Verbena / Root (washed) / Verbena / Verbenaceae Verbeneae Verbena |  |
||
| Plant;Root | Dianthus Olivia / Root (washed) / Dianthus / Caryophyllaceae Dianthus |  |
||
| Plant;Root | Dianthus / Root (washed) / Dianthus / Caryophyllaceae Dianthus |  |
||
| Plant;Root | Scarlet sage / Root (washed) / Salvia splendens / Lamiaceae Nepetoideae Mentheae Salvia |  |
||
| Herbal medicine;Plant | Bakumondoto (Extract Granules) |  |
||
| Herbal medicine;Plant | Shimbuto (Extract Granules) |  |
||
| Herbal medicine;Plant | Goshuyuto (Extract Granules) |  |
||
| Herbal medicine;Plant | Ninjinto (Extract Granules) |  |
||
| Plant;Shoot | Parsley / Shoot / Petroselinum crispum / Apiaceae Petroselinum |  |
||
| Plant;Shoot | Rosemary (crawling) / Shoot / Rosmarinus officinalis L. / Lamiaceae Rosmarinus |  |
||
| Plant;Shoot | Mint / Shoot / Mentha sp. / Lamiaceae Mentha |  |
||
| Plant;Shoot | Lemon balm / Shoot / Melissa officinalis / Lamiaceae Melissa |  |
||
| Plant;Shoot | Basil / Shoot / Ocimum basilicum / Lamiaceae Nepetoideae Ocimeae Ocimum |  |
||
| Plant;Flower | Hop dried flower / Humulus lupulus / Cannabaceae Humulus |  |
||
| Plant;Seed | Coffee bean, roasted, arabica / Coffea arabica L. / Rubiaceae Coffea |  |
||
| Plant;Seed | Coffee bean, roasted, robusta / Coffea canephora / Rubiaceae Coffea |  |
||
| Food;Plant | Coffee, infusion, arabica / Coffea arabica L. / Rubiaceae Coffea |  |
||
| Food;Plant | Coffee, infusion, robusta / Coffea canephora / Rubiaceae Coffea |  |
||
| Herbal medicine;Plant | Daiobotampito (Extract Granules) |  |
||
| Herbal medicine;Plant | Byakkokaninjinto (Extract Granules) |  |
||
| Herbal medicine;Plant | Shigyakusan (Extract Granules) |  |
||
| Herbal medicine;Plant | Mokuboito (Extract Granules) |  |
||
| Plant;Seed | Coffee bean, unroasted, arabica / Coffea arabica L. / Rubiaceae Coffea |  |
||
| Plant;Seed | Coffee bean, unroasted, robusta / Coffea canephora / Rubiaceae Coffea |  |
||
| Plant;Flower | Fuchsia / Flower, Flower bud / Fuchsia L. / Onagraceae Fuchsia |  |
||
| Plant;Shoot | Fuchsia / Leaf, Stem, Flower bud (small) / Fuchsia L. / Onagraceae Fuchsia |  |
||
| Plant;Shoot | Bitter melon / Leaf, Stem, Vine / Momordica charantia / Cucurbitaceae Momordica |  |
||
| Plant;Shoot | Melon / Leaf, Stem / Cucumis melo / Cucurbitaceae Cucumis |  |
||
| Plant;Shoot | Watermelon / Leaf, Stem / Citrullus lanatus / Cucurbitaceae Citrullus |  |
||
| Plant;Shoot | Cucumber 1 / Leaf, Stem, Vine / Cucumis sativus L. / Cucurbitaceae Cucumis |  |
||
| Plant;Shoot | Cucumber 2 / Leaf, Stem, Vine / Cucumis sativus L. / Cucurbitaceae Cucumis |  |
||
| Plant;Shoot | Perilla (Ao-shiso) / Leaf, Stem / Perilla frutescens var. crispa / Lamiaceae Perilla |  |
||
| Plant;Shoot | Eggplant / Leaf and Petiole (excluding the part of feeding damage) / Solanum melongena / Solanaceae Solanum |  |
||
| Animal;Urine | Dog / Urine / Canis lupus familiaris / Canidae Caninae Canis |  |
||
| Animal;Urine | Cat / Urine / Felis silvestris catus / Felidae Felis |  |
||
| Product | Dry dog food |  |
||
| Plant;Shoot | Luffa / Leaf, Stem, Vine / Luffa cylindrica / Cucurbitaceae Luffa |  |
||
| Plant;Shoot | Tomato / Leaf, Stem / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Shoot | Shishito pepper / Leaf and Stem (a bit damaged by feeding) / Capsicum annuum / Solanaceae Capsicum |  |
||
| Plant;Shoot | Red pepper / Leaf, Stem / Capsicum annuum / Solanaceae Capsicum |  |
||
| Plant;Shoot | Green pepper / Leaf, Stem / Capsicum annuum / Solanaceae Capsicum |  |
||
| Plant | Fava bean / Shell / Vicia faba / Fabaceae Vicia |  |
||
| Plant;Seed;Food | Fava bean / Bean / Vicia faba / Fabaceae Vicia |  |
||
| Plant;Flower | Sanvitalia Greatyellow / Flower / Sanvitalia / Asteraceae Sanvitalia |  |
||
| Plant;Shoot | Sanvitalia Greatyellow / Leaf, Stem, Flower bud (small) / Sanvitalia / Asteraceae Sanvitalia |  |
||
| Plant;Shoot | Japanese pepper / Leaf, Stem (branch) / Zanthoxylum piperitum / Rutaceae Zanthoxylum |  |
||
| Microbes;Yeast | Fission yeast L968 (NBRP FY7520) / Spores (formed 6 days on synthetic medium) / Schizosaccharomyces pombe / Schizosaccharomycetaceae Schizosaccharomyces |  |
||
| Medium | Synthetic medium (for spore formation, used for 6 days, NBRP) |  |
||
| Medium | Synthetic medium (for spore formation, unused, NBRP) |  |
||
| Microbes;Yeast | Fission yeast L968 (NBRP FY7520) / Spores (formed 6 days on wheat germ medium) / Schizosaccharomyces pombe / Schizosaccharomycetaceae Schizosaccharomyces |  |
||
| Medium | Wheat germ medium (for spore formation, used for 6 days, NBRP) |  |
||
| Medium | Wheat germ medium (for spore formation, unused, NBRP) |  |
||
| Plant;Seed | Barley (Morex) NBRP / Seed / Hordeum vulgare subsp. vulgare cv. Morex / Poaceae Hordeum |  |
||
| Plant;Seed | Barley (Haruna Nijo) NBRP / Seed / Hordeum vulgare subsp. vulgare cv. Haruna Nijo / Poaceae Hordeum |  |
||
| Plant;Seed | Barley (Murasakimochi) NBRP / Seed / Hordeum vulgare subsp. vulgare cv. Murasakimochi / Poaceae Hordeum |  |
||
| Plant;Seed | Barley (Golden Promise) NBRP / Seed / Hordeum vulgare subsp. vulgare cv. Golden Promise / Poaceae Hordeum |  |
||
| Plant;Seed | Barley (wild strain OUH602) NBRP / Seed / Hordeum vulgare subsp. spontaneum, strain OUH602 / Poaceae Hordeum |  |
||
| Herbal medicine;Plant | Hangebyakujutsutemmato (Extract Granules) |  |
||
| Herbal medicine;Plant | Tokishigyakukagoshuyushokyoto (Extract Granules) |  |
||
| Herbal medicine;Plant | Ryokeijutsukanto (Extract Granules) |  |
||
| Herbal medicine;Plant | Choreito (Extract Granules) |  |
||
| Herbal medicine;Plant | Hochuekkito (Extract Granules) |  |
||
| Herbal medicine;Plant | Rikkunshito (Extract Granules) |  |
||
| Herbal medicine;Plant | Keishito (Extract Granules) |  |
||
| Herbal medicine;Plant | Shichimotsukokato (Extract Granules) |  |
||
| Plant;Root | Perilla (Ao-shiso) / Root (washed) / Perilla frutescens var. crispa / Lamiaceae Perilla |  |
||
| Plant;Root | Eggplant / Root (washed) / Solanum melongena / Solanaceae Solanum |  |
||
| Plant;Root | Cucumber 1 / Root (washed) / Cucumis sativus L. / Cucurbitaceae Cucumis |  |
||
| Plant;Root | Cucumber 2 / Root (washed) / Cucumis sativus L. / Cucurbitaceae Cucumis |  |
||
| Plant;Root | Sanvitalia Greatyellow / Root (washed) / Sanvitalia / Asteraceae Sanvitalia |  |
||
| Plant;Root | Luffa / Root (washed) / Luffa cylindrica / Cucurbitaceae Luffa |  |
||
| Plant;Root | Melon / Root (washed) / Cucumis melo / Cucurbitaceae Cucumis |  |
||
| Plant;Root | Tomato / Root (washed) / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Root | Bitter melon / Root (washed) / Momordica charantia / Cucurbitaceae Momordica |  |
||
| Plant;Root | Fuchsia / Root (washed) / Fuchsia L. / Onagraceae Fuchsia |  |
||
| Plant;Root | Watermelon / Root (washed) / Citrullus lanatus / Cucurbitaceae Citrullus |  |
||
| Microbes;Yeast | Fission yeast L972 (NBRP FY7507) / Cell (grown in synthetic medium with NH4+ as nitrogen source) / Schizosaccharomyces pombe / Schizosaccharomycetaceae Schizosaccharomyces |  |
||
| Microbes;Yeast | Fission yeast L972 (NBRP FY7507) / Cell (grown in minimal medium with proline as nitrogen source) / Schizosaccharomyces pombe / Schizosaccharomycetaceae Schizosaccharomyces | 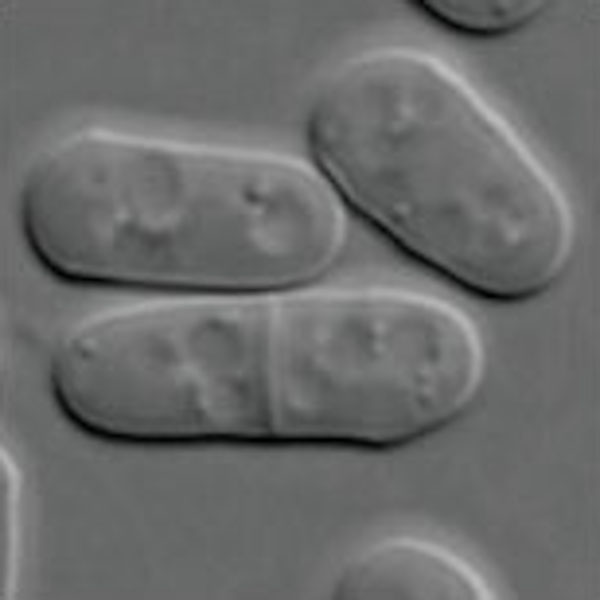 |
||
| Medium | Minimal medium with proline as nitrogen source (for fission yeast, used, NBRP) |  |
||
| Medium | Minimal medium with proline as nitrogen source (for fission yeast, unused, NBRP) |  |
||
| Food;Plant;Fruit | Grape 'Shine Muscat' / Fruit / Vitis / Vitaceae Vitis |  |
||
| Food;Plant;Fruit | Grape 'Tanenashi Kyoho' / Fruit / Vitis / Vitaceae Vitis |  |
||
| Food;Plant;Fruit | Grape 'Kaiji' / Fruit / Vitis / Vitaceae Vitis |  |
||
| Food;Plant;Fruit | Olive, black olive, flavored / / Olea europaea / Oleaceae Olea |  |
||
| Food;Plant | Grape juice, from concentrate, 100% / Vitis / Vitaceae Vitis |  |
||
| Plant;Flower | Morning glory (NBRP Q1065, synonym AK001) / Flower / Ipomoea nil / Convolvulaceae Convolvuloideae Ipomoneeae Ipomoea |  |
||
| Plant;Shoot | Morning glory (NBRP Q1065, synonym AK001) / Stem / Ipomoea nil / Convolvulaceae Convolvuloideae Ipomoneeae Ipomoea |  |
||
| Plant;Leaf | Morning glory (NBRP Q1065, synonym AK001) / Leaf / Ipomoea nil / Convolvulaceae Convolvuloideae Ipomoneeae Ipomoea |  |
||
| Plant;Root | Morning glory (NBRP Q1065, synonym AK001) / Root / Ipomoea nil / Convolvulaceae Convolvuloideae Ipomoneeae Ipomoea |  |
||
| Plant;Seed | Morning glory (NBRP Q1065, synonym AK001) / Seed / Ipomoea nil / Convolvulaceae Convolvuloideae Ipomoneeae Ipomoea |  |
||
| Plant;Leaf | Tomato Micro-Tom (NBRP TOMJPF00001) / Leaf / Solanum lycopersicum / Solanaceae Slanum |  |
||
| Plant;Root | Tomato Micro-Tom (NBRP TOMJPF00001) / Root / Solanum lycopersicum / Solanaceae Slanum |  |
||
| Plant;Leaf | Tomato Moneymaker (NBRP TOMJPF00002) / Leaf / Solanum lycopersicum / Solanaceae Slanum |  |
||
| Plant;Root | Tomato Moneymaker (NBRP TOMJPF00002) / Root / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Leaf | Tomato Aichi First (NBRP TOMJPF00003) / Leaf / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Root | Tomato Aichi First (NBRP TOMJPF00003) / Root / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Leaf | Tomato Ailsa Craig (NBRP TOMJPF00004) / Leaf / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Root | Tomato Ailsa Craig (NBRP TOMJPF00004) / Root / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Leaf | Tomato M82 (NBRP TOMJPF00005) / Leaf / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Root | Tomato M82 (NBRP TOMJPF00005) / Root / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Leaf | Tomato Rutgers (NBRP TOMJPF00006) / Leaf / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Root | Tomato Rutgers (NBRP TOMJPF00006) / Root / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Leaf | Tomato Ponderosa (NBRP TOMJPF00007) / Leaf / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Root | Tomato Ponderosa (NBRP TOMJPF00007) / Root / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Leaf | Tomato Ueleie106WP (NBRP TOMJPF00015) / Leaf / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Root | Tomato Ueleie106WP (NBRP TOMJPF00015) / Root / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Leaf | Tomato Heinz1706B (NBRP TOMJPF00016) / Leaf / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Root | Tomato Heinz1706B (NBRP TOMJPF00016) / Root / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Leaf | Tomato Cerasiforme (NBRP TOMJPF00009) / Leaf / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Root | Tomato Cerasiforme (NBRP TOMJPF00009) / Root / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Leaf | Tomato S. peruvianum 1 (NBRP TOMJPF00010-1) / Leaf / Solanum peruvianum / Solanaceae Solanum |  |
||
| Plant;Root | Tomato S. peruvianum 1 (NBRP TOMJPF00010-1) / Root / Solanum peruvianum / Solanaceae Solanum | 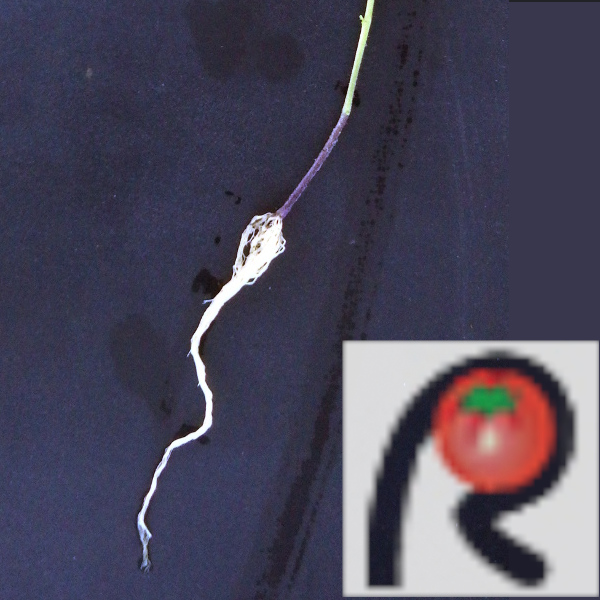 |
||
| Plant;Leaf | Tomato S. peruvianum 2 (NBRP TOMJPF00010-2) / Leaf / Solanum peruvianum / Solanaceae Solanum |  |
||
| Plant;Root | Tomato S. peruvianum 2 (NBRP TOMJPF00010-2) / Root / Solanum peruvianum / Solanaceae Solanum |  |
||
| Plant;Leaf | Tomato S. pimpinellifolium 1 (NBRP TOMJPF00012-1) / Leaf / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Root | Tomato S. pimpinellifolium 1 (NBRP TOMJPF00012-1) / Root / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Leaf | Tomato S. pimpinellifolium 2 (NBRP TOMJPF00012-2) / Leaf / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Root | Tomato S. pimpinellifolium 2 (NBRP TOMJPF00012-2) / Root / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Leaf | Tomato S. pimpinellifolium 3 (NBRP TOMJPF00012-3) / Leaf / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Root | Tomato S. pimpinellifolium 3 (NBRP TOMJPF00012-3) / Root / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Leaf | Tomato S. pimpinellifolium 4 (NBRP TOMJPF00012-4) / Leaf / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Root | Tomato S. pimpinellifolium 4 (NBRP TOMJPF00012-4) / Root / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Leaf | Tomato S. pennelii (NBRPTOMJPF00008) / Leaf / Solanum pennellii / Solanaceae Solanum |  |
||
| Plant;Leaf | Tomato S. peruvianum 3 (NBRP TOMJPF00010-3) / Leaf / Solanum peruvianum / Solanaceae Solanum |  |
||
| Plant;Root | Parsley / Root (washed) / Petroselinum crispum / Apiaceae Petroselinum |  |
||
| Plant;Root | Red pepper / Root (washed) / Capsicum annuum / Solanaceae Capsicum |  |
||
| Plant;Root | Shishito pepper / Root (washed) / Capsicum annuum / Solanaceae Capsicum |  |
||
| Plant;Root | Green pepper / Root (washed) / Capsicum annuum / Solanaceae Capsicum |  |
||
| Plant;Root | Lemon balm / Root (washed) / Melissa officinalis / Lamiaceae Melissa |  |
||
| Plant;Root | Mint / Root (washed) / Mentha sp. / Lamiaceae Mentha |  |
||
| Plant;Root | Basil / Root (washed) / Ocimum basilicum / Lamiaceae Nepetoideae Ocimeae Ocimum |  |
||
| Plant;Root | Rosemary (crawling) / Root (washed) / Rosmarinus officinalis L. / Lamiaceae Rosmarinus |  |
||
| Food;Plant;Seed | Paddy rice 'Akitakomachi' (non-glutinous) / Grain, raw (well-milled, wash-free) / Oryza sativa L. / Poaceae Oryzoideae Oryza |  |
||
| Food;Plant;Seed | Paddy rice 'Akitakomachi' (non-glutinous) / Grain, raw (brown rice) / Oryza sativa L. / Poaceae Oryzoideae Oryza |  |
||
| Food;Plant;Seed | Barley / Grain, raw (rolled barley) / Hordeum vulgare / Poaceae Hordeum |  |
||
| Food;Plant;Seed | Paddy rice 'Akitakomachi' (non-glutinous) / Cooked rice (well-milled, wash-free) / Oryza sativa L. / Poaceae Oryzoideae Oryza |  |
||
| Food;Plant;Seed | Paddy rice 'Akitakomachi' (non-glutinous) / Cooked rice (brown rice) / Oryza sativa L. / Poaceae Oryzoideae Oryza |  |
||
| Food;Plant;Seed | Barley / Cooked grain (rolled barley) / Hordeum vulgare / Poaceae Hordeum |  |
||
| Food;Animal | Domestic fowl / Hen's egg (Yolk and White) / Gallus gallus / Phasianidae Phasianoidea Gallus |  |
||
| Food;Plant | Wakame seaweed / Water reconstituted from dried seaweed / Undaria pinnatifida / Alariaceae Undaria |  |
||
| Food;Plant | Green tea (sencha) / Infusion / Camellia sinensis / Theaceae Camellia |  |
||
| Food;Plant;Seed | Soybean / Seed, mature, raw / Glycine max / Fabaceae Faboideae Glycine |  |
||
| Food;Plant;Seed | Sesame / Seed (roasted sesame) / Sesamum indicum / Pedaliaceae Sesamum |  |
||
| Food;Plant | Green tea (sencha) / Tea leaf / Camellia sinensis / Theaceae Camellia |  |
||
| Food;Microbes;Fungi | Shiitake mushroom / edible part, raw / Lentinula edodes / Tricholomataceae Lentinula |  |
||
| Food;Plant;Fruit | Satsuma mandarin / Segment / Citrus unshiu / Rutaceae Citrus |  |
||
| Food;Plant;Fruit | Satsuma mandarin / Segment (with membrane) / Citrus unshiu / Rutaceae Citrus |  |
||
| Plant | Satsuma mandarin / Peel (with hull, washed) / Citrus unshiu / Rutaceae Citrus |  |
||
| Food;Plant;Fruit | Apple / Edible part (with peel, washed) / Malus domestica / Rosaceae Amygdaloideae Malus |  |
||
| Plant;Seed | Apple / Seed / Malus domestica / Rosaceae Amygdaloideae Malus |  |
||
| Food;Plant;Fruit | Squash / Edible part (with peel, washed) / Cucurbita L. / Cucurbitaceae Cucurbiteae Cucurbita |  |
||
| Plant;Seed | Squash / Seed, raw / Cucurbita L. / Cucurbitaceae Cucurbiteae Cucurbita |  |
||
| Food;Plant | Onion / Bulb, raw / Allium cepa / Amaryllidaceae Allioideae Allium |  |
||
| Food;Plant;Fruit | Eggplant / Fruit, raw (edible part, with peel, washed) / Solanum melongena / Solanaceae Solanum |  |
||
| Food;Plant;Root | Carrot / Tuberous root, raw (edible part, with peel, washed) / Daucus carota subsp. sativus / Apiaceae Daucus |  |
||
| Food;Plant;Root | Potato / Tuber, raw (edible part, without peel) / Solanum tuberosum / Solanaceae Solanum |  |
||
| Food;Plant;Leaf | Cabbage / Leaf (edible part) / Brassica oleracea var. capitata / Brassicaceae Brassica |  |
||
| Food;Plant;Root | Sweet potato / Tuberous root, raw (edible part, with peel, washed) / Ipomoea batatas / Convolvulaceae Ipomoea |  |
||
| Food;Plant;Root | Burdock / Tuberous root, raw (edible part, without peel) / Arctium lappa / Asteraceae Arctium |  |
||
| Food;Plant;Root | Burdock / Tuberous root, raw (edible part, with peel, washed) / Arctium lappa / Asteraceae Arctium |  |
||
| Food;Plant;Fruit | Green pepper / Fruit, raw (edible part, washed) / Capsicum annuum / Solanaceae Capsicum |  |
||
| Food;Plant;Fruit | Green pepper / Core and seed / Capsicum annuum / Solanaceae Capsicum |  |
||
| Food;Plant;Root | Japanese radish (Daikon) / Tuberous root, raw (edible part, with peel, washed) / Raphanus sativus var. hortensis / Brassicaceae Raphanus |  |
||
| Food;Plant;Leaf | Welsh onion / Leaf, raw (edible part, washed) / Allium fistulosum / Amaryllidaceae Allioideae Allium |  |
||
| Food;Plant;Leaf | Spinach mustard (Komatsuna) / Leaf, raw (edible part, washed) / Brassica rapa var. perviridis / Brassicaceae Brassica |  |
||
| Food;Plant;Leaf | Spinach / Leaf, raw (edible part, washed) / Spinacia oleracea / Amaranthaceae Chenopodioideae Spinacia |  |
||
| Food;Plant;Leaf | Spinach / Base, raw (washed) / Spinacia oleracea / Amaranthaceae Chenopodioideae Spinacia |  |
||
| Food;Animal | Domestic fowl / Breast meat / Gallus gallus / Phasianidae Phasianoidea Gallus |  |
||
| Herbal medicine;Plant | Chotosan (Extract Granules) |  |
||
| Herbal medicine;Plant | Juzentaihoto (Extract Granules) |  |
||
| Herbal medicine;Plant | Keigairengyoto (Extract Granules) |  |
||
| Herbal medicine;Plant | Junchoto (Extract Granules) |  |
||
| Herbal medicine;Plant | Yokuininto (Extract Granules) |  |
||
| Herbal medicine;Plant | Sokeikakketsuto (Extract Granules) |  |
||
| Herbal medicine;Plant | Yokukansan (Extract Granules) |  |
||
| Herbal medicine;Plant | Makyokansekito (Extract Granules) |  |
||
| Herbal medicine;Plant | Gorinsan (Extract Granules) |  |
||
| Herbal medicine;Plant | Unseiin (Extract Granules) |  |
||
| Herbal medicine;Plant | Seijobofuto (Extract Granules) |  |
||
| Herbal medicine;Plant | Jizusoippo (Extract Granules) |  |
||
| Herbal medicine;Plant | Keishikashakuyakuto (Extract Granules) |  |
||
| Herbal medicine;Plant | Tokakujokito (Extract Granules) |  |
||
| Herbal medicine;Plant | Bofutsushosan (Extract Granules) |  |
||
| Herbal medicine;Plant | Goshakusan (Extract Granules) |  |
||
| Plant;Leaf | Camphor Laurel / Leaf / Cinnamomum camphora / Lauraceae Cinnamomum |  |
||
| Plant:Shoot | Round leaf holly / Leaf, Stem / Ilex rotunda / Aquifoliaceae Ilex |  |
||
| Plant:Shoot | Japanese hackberry / Leaf, Stem / Celtis sinensis / Cannabaceae Celtis |  |
||
| Plant:Shoot | Heavenly bamboo / Leaf / Nandina domestica / Berberidaceae Nandinoideae Nandina |  |
||
| Plant;Flower | Heavenly bamboo / Flower / Nandina domestica / Berberidaceae Nandinoideae Nandina |  |
||
| Plant:Shoot | Japanese wisteria / Leaf, Stem / Wisteria floribunda / Fabaceae Faboideae Millettieae Wisteria |  |
||
| Plant;Leaf | Japanese privet / Leaf / Ligustrum japonicum / Oleaceae Ligustrum |  |
||
| Plant;Flower | Japanese privet / Flower / Ligustrum japonicum / Oleaceae Ligustrum |  |
||
| Plant;Leaf | Persimmon / Leaf / Diospyros kaki Thunb. / Ebenaceae Diospyros |  |
||
| Plant:Shoot | Tree of a thousand stars / Leaf, Stem (young), Flower / Serissa japonica / Rubiaceae Rubioideae Paederieae Serissa |  |
||
| Plant;Leaf | Japanese wild raspberry / Leaf / Rubus trifidus / Rosaceae Rosoideae Rubus |  |
||
| Plant:Shoot | Japanese holly / Leaf, Stem / Ilex crenata / Aquifoliaceae Ilex |  |
||
| Plant;Leaf | Hama-giku (a kind of chrysanthemum, NBRP YPK35) / Leaf / Nipponanthemum nipponicum (Franch. ex Maxim.) Kitamura / Asteraceae Asteroideae Anthemideae Nipponanthemum |  |
||
| Plant;Flower | Hama-giku (a kind of chrysanthemum) / Flower / Nipponanthemum nipponicum (Franch. ex Maxim.) Kitamura / Asteraceae Asteroideae Anthemideae Nipponanthemum |  |
||
| Plant;Leaf | Kawarayomogi (a kind of artemisia, NBRP KZ01) / Leaf / Artemisia capillaris Thunb. / Asteraceae Asteroideae Artemisia |  |
||
| Plant;Flower | Kawarayomogi (a kind of artemisia, NBRP KZ01) / Flower / Artemisia capillaris Thunb. / Asteraceae Asteroideae Artemisia |  |
||
| Plant;Leaf | Inuyomogi (a kind of artemisia, NBRP X2NW01) / Leaf / Artemisia keiskeana Miq. / Asteraceae Asteroideae Artemisia |  |
||
| Plant;Flower | Inuyomogi (a kind of artemisia, NBRP X2NW01) / Flower / Artemisia keiskeana Miq. / Asteraceae Asteroideae Artemisia |  |
||
| Plant;Leaf | Ryuno-giku (a kind of chrysanthemum, NBRP ABP23) / Leaf / Chrysanthemum makinoi Matsum. et Nakai / Asteraceae Chrysanthemum |  |
||
| Plant;Flower | Ryuno-giku (a kind of chrysanthemum, NBRP ABP23) / Flower / Chrysanthemum makinoi Matsum. et Nakai / Asteraceae Chrysanthemum |  |
||
| Plant;Leaf | Kikutani-giku (a kind of chrysanthemum, NBRP AEV12) / Leaf / Chrysanthemum seticuspe (Maxim.) Hand.-Mazz. / Asteraceae Chrysanthemum |  |
||
| Plant;Flower | Kikutani-giku (a kind of chrysanthemum, NBRP AEV12) / Flower / Chrysanthemum seticuspe (Maxim.) Hand.-Mazz. / Asteraceae Chrysanthemum |  |
||
| Standard Chemical | (+/-)-Anabasine |  |
||
| Herbal medicine;Plant | Shakanzoto (Extract Granules) |  |
||
| Herbal medicine;Plant | Kihito (Extract Granules) |  |
||
| Herbal medicine;Plant | Jinsoin (Extract Granules) |  |
||
| Herbal medicine;Plant | Nyoshinsan (Extract Granules) |  |
||
| Herbal medicine;Plant | Shakuyakukanzoto (Extract Granules) |  |
||
| Herbal medicine;Plant | Bukuryoin (Extract Granules) |  |
||
| Herbal medicine;Plant | Kososan (Extract Granules) |  |
||
| Herbal medicine;Plant | Shimotsuto (Extract Granules) |  |
||
| Herbal medicine;Plant | Kambakutaisoto (Extract Granules) |  |
||
| Herbal medicine;Plant | Saikanto (Extract Granules) |  |
||
| Herbal medicine;Plant | Choijokito (Extract Granules) |  |
||
| Herbal medicine;Plant | Shikunshito (Extract Granules) |  |
||
| Herbal medicine;Plant | Ryutanshakanto (Extract Granules) |  |
||
| Herbal medicine;Plant | Kyukikyogaito (Extract Granules) |  |
||
| Herbal medicine;Plant | Makyoyokukanto (Extract Granules) |  |
||
| Herbal medicine;Plant | Heiisan (Extract Granules) |  |
||
| Herbal medicine;Plant | Saikoseikanto (Extract Granules) |  |
||
| Herbal medicine;Plant | Nichinto (Extract Granules) |  |
||
| Herbal medicine;Plant | Keishininjinto (Extract Granules) |  |
||
| Herbal medicine;Plant | Yokukansankachimpihange (Extract Granules) |  |
||
| Herbal medicine;Plant | Daiokanzoto (Extract Granules) |  |
||
| Herbal medicine;Plant | Shimpito (Extract Granules) |  |
||
| Herbal medicine;Plant | Tokiinshi (Extract Granules) |  |
||
| Herbal medicine;Plant | Rokumigan (Extract Granules) |  |
||
| Plant:Shoot | Bushkiller / Leaf, Stem / Cayratia japonica / Vitaceae Cayratia |  |
||
| Plant;Leaf | Japanese silver leaf / Leaf / Farfugium japonicum / Asteraceae Asteroideae Farfugium |  |
||
| Plant;Leaf | Spiderwort / Leaf / Tradescantia ohiensis / Commelinaceae Tradescantia |  |
||
| Plant;Flower | Spiderwort / Flower, Flower bud / Tradescantia ohiensis / Commelinaceae Tradescantia |  |
||
| Plant:Shoot | A kind of Firethorn (putative) / Leaf, Stem / Pyracantha sp. (putative) / Rosaceae Maloideae Pyracantha |  |
||
| Plant;Leaf | Chinese holly / Leaf / Osmanthus heterophyllus / Oleaceae Osmanthus |  |
||
| Plant;Leaf | Tiger lily / Leaf / Lilium lancifolium / Liliaceae Lilium |  |
||
| Plant;Leaf | Sawara cypress / Leaf / Chamaecyparis pisifera / Cupressaceae Chamaecyparis |  |
||
| Plant;Leaf | Camellia / Leaf / Camellia japonica / Theaceae Camellia |  |
||
| Plant;Leaf | Japanese aralia / Leaf / Fatsia japonica / Araliaceae Fatsia |  |
||
| Plant:Shoot | Japanese creeping juniper (putative) / Leaf, Stem / Juniperus procumbens (putative) / Cupressaceae Juniperus |  |
||
| Plant:Shoot | A kind of Stonecrop (putative) / Leaf, Stem, Flower / Sedum japonicum subsp. oryzifolium (putative) / Crassulaceae Sempervivoideae Sedeae Sedinae Sedum |  |
||
| Plant;Leaf | Japanese apricot / Leaf / Prunus mume / Rosaceae Prunus |  |
||
| Plant;Leaf | Flowering peach / Leaf / Prunus persica / Rosaceae Prunus |  |
||
| Plant;Leaf | Sasanqua / Leaf / Camellia sasanqua / Theaceae Camellia |  |
||
| Plant;Leaf | Flowering quince / Leaf / Chaenomeles speciosa / Rosaceae Amygdaloideae Maleae Chaenomeles |  |
||
| Plant;Leaf | Rose / Leaf / Rosa sp. / Rosaceae Rosoideae Rosa |  |
||
| Plant;Leaf | Japanese rose / Leaf / Rosa multiflora / Rosaceae Rosoideae Rosa |  |
||
| Plant;Leaf | Japanese Aucuba / Leaf / Aucuba japonica / Garryaceae Aucuba |  |
||
| Plant;Leaf | Japanese kerria / Leaf / Kerria japonica / Rosaceae Amygdaloideae Kerria |  |
||
| Plant;Leaf | Hellebore / Leaf / Helleborus sp. / Ranunculaceae Ranunculoideae Helleboreae Helleborus |  |
||
| Plant;Leaf | Chrysanthemum / Leaf / Chrysanthemum morifolium / Asteraceae Chrysanthemum |  |
||
| Plant:Shoot | Pomegranate / Leaf, Stem / Punica granatum / Lythraceae Punica |  |
||
| Plant;Leaf | German iris / Leaf / Iris germanica / Iridaceae Iris |  |
||
| Plant;Leaf | Siberian iris / Leaf / Iris sanguinea / Iridaceae Iris |  |
||
| Plant;Leaf | Chinese redbud / Leaf / Cercis chinensis / Fabaceae Caesalpinioideae Cercis |  |
||
| Plant;Fruit | Chinese redbud / Pod / Cercis chinensis / Fabaceae Caesalpinioideae Cercis |  |
||
| Plant;Leaf | Blueberry / Leaf / Vaccinium sp. / Ericaceae Vaccinoideae Vaccinium |  |
||
| Plant;Fruit | Blueberry / Fruit (young) / Vaccinium sp. / Ericaceae Vaccinoideae Vaccinium |  |
||
| Plant;Leaf | Flowering dogwood / Leaf / Cornus florida / Cornaceae Cornus |  |
||
| Plant;Leaf | Kumquat / Leaf / Citrus japonica / Rutaceae Citrus |  |
||
| Plant;Leaf | Pomegranate / Leaf / Punica granatum / Lythraceae Punica |  |
||
| Plant;Flower | Pomegranate / Flower, Flower bud / Punica granatum / Lythraceae Punica |  |
||
| Plant;Leaf | Mountain hydrangea / Leaf / Hydrangea serrata / Hydrangeaceae Hydrangea |  |
||
| Plant;Leaf | Gooseberry / Leaf / Ribes uva-crispa / Grossulariaceae Ribes |  |
||
| Plant;Fruit | Gooseberry / Fruit (immature) / Ribes uva-crispa / Grossulariaceae Ribes |  |
||
| Plant;Leaf | Cotton rose / Leaf / Hibiscus mutabilis cv. Versicolor / Malvaceae Malvoideae Hibisceae Hibiscus |  |
||
| Plant:Shoot | Cootamundra wattle / Leaf, Stem (young) / Acacia baileyana / Fabaceae Mimosoideae Acacia |  |
||
| Plant;Leaf | Persimmon / Leaf / Diospyros kaki Thunb. / Ebenaceae Diospyros |  |
||
| Plant;Leaf | Nagi, Asian bayberry / Leaf / Nageia nagi / Podocarpaceae Nageia |  |
||
| Plant:Shoot | Japanese eurya / Leaf, Stem (young) / Eurya japonica / Pentaphylacaceae Eurya |  |
||
| Plant;Leaf | Hyacinth orchid / Leaf / Bletilla striata / Orchidaceae Epidendroideae Arethuseae Bletilla |  |
||
| Plant;Leaf | Fragrant orange-colored olive / Leaf / Osmanthus fragrans var. aurantiacus / Oleaceae Oleeae Osmanthus |  |
||
| Plant;Leaf | Downy cherry / Leaf / Prunus tomentosa / Rosaceae Prunus |  |
||
| Plant;Fruit | Downy cherry / Fruit / Prunus tomentosa / Rosaceae Prunus |  |
||
| Standard Chemical | Standard Nicotinic acid |  |
||
| Standard Chemical | Standard Nicotinamide |  |
||
| Standard Chemical | Standard Isonicotinamide |  |
||
| Plant:Shoot | Winter daphne / Leaf, Stem (young) / Daphne odora / Thymelaeaceae Daphne |  |
||
| Plant;Leaf | Maple / Leaf / Acer sp. / Sapindaceae Acer |  |
||
| Plant;Leaf | Navel orange / Leaf / Citrus sinensis / Rataceae Citrus |  |
||
| Plant;Leaf | Magnolia / Leaf / Magnolia liliiflora / Magnoliaceae Magnolia |  |
||
| Plant;Leaf | Japanese cinnamon tree / Leaf / Cinnamomum sieboldii / Laureaceae Cinnamomum |  |
||
| Plant;Leaf | Japanese star anse (putative) / Leaf / Illicium anisatum / Schisandraceae Illicium |  |
||
| Plant;Leaf | Potato / Leaf / Solanum tuberosum / Solanaceae Solanum |  |
||
| Plant;Flower | Prairie fleabane / Flower, Flower bud / Erigeron strigosus / Asteraceae Asteroideae Astereae Erigeron |  |
||
| Plant:Shoot | Prairie fleabane / Leaf, Stem / Erigeron strigosus / Asteraceae Asteroideae Astereae Erigeron |  |
||
| Plant;Flower | Dokudami / Flower, Flower bud / Houttuynia cordata / Saururaceae Houttuynia |  |
||
| Plant:Shoot | Dokudami / Leaf, Stem / Houttuynia cordata / Saururaceae Houttuynia |  |
||
| Herbal medicine;Plant | Nijutsuto (Extract Granules) |  |
||
| Plant;Flower | Kousa Dogwood / Flower / Cornus kousa / Cornaceae Cornus |  |
||
| Plant;Leaf | Kousa Dogwood / Leaf / Cornus kousa / Cornaceae Cornus |  |
||
| Plant;Leaf | Southern catalpa / Leaf / Catalpa bignonioides Walt. / Bignoniaceae Catalpa |  |
||
| Plant;Flower | Southern catalpa / Flower, Flower bud / Catalpa bignonioides Walt. / Bignoniaceae Catalpa |  |
||
| Plant;Leaf | Japanese chinquapin / Leaf / Castanopsis cuspidata Schottky / Fagaceae Castanopsis |  |
||
| Plant;Leaf | Quercus phylliraeoides / Leaf / Quercus phylliraeoides A. Gray / Fagaceae Quercus |  |
||
| Plant;Leaf | Kobushi magnolia / Leaf / Magnolia kobus DC. / Magnoliaceae Magnolia |  |
||
| Plant:Shoot | Japanese holly / Leaf, Stem (young) / Ilex crenata Thunb. / Aquifoliaceae Ilex |  |
||
| Plant;Leaf | Japanese cleyera / Leaf (young) / Ternstroemia gymnanthera Beddone. / Pentaphylacaceae Ternstroemia |  |
||
| Plant;Leaf | Tall Stewartia / Leaf / Stewartia monadelpha / Theaceae Stewartia |  |
||
| Plant;Leaf | Threadleaf false cypress / Leaf (young) / Chamaecyparis pisifera Endl. cv. filifera / Cupressaceae Cupressoideae Chamaecyparis |  |
||
| Plant;Leaf | Red bayberry / Leaf (young) / Myrica rubra Sieb. et Zucc. / Myricaceae Myrica |  |
||
| Plant;Leaf | Southern magnolia / Leaf (young) / Magnolia grandiflora Linn. / Magnoliaceae Magnolia |  |
||
| Plant;Leaf | Himalayan cedar / Leaf / Cedrus deodara Loud. / Pinaceae Cedrus |  |
||
| Plant;Leaf | Round leaf holly / Leaf (young) / Ilex rotunda Thunb. / Aquifoliaceae Ilex |  |
||
| Plant;Leaf | Pokeweed / Leaf / Phytolacca americana / Phytolaccaceae Phytolacca |  |
||
| Plant;Fruit | Pokeweed / Fruit (immature) / Phytolacca americana / Phytolaccaceae Phytolacca |  |
||
| Plant;Leaf | Harlequin glorybower (putative) / Leaf / Clerodendrum trichotomum (putative) / Lamiaceae Ajugoideae Clerodendrum |  |
||
| Plant;Fruit | Wild Korean mulberry (putative) / Fruit / Morus australis (putative) / Moraceae Morus |  |
||
| Plant;Leaf | An water-lily species. / Leaf (floating leaf) / Nymphaea sp. / Nymphaeaceae Nymphaea |  |
||
| Plant;Leaf | Tall Stewartia / Leaf / Stewartia monadelpha / Theaceae Stewartia |  |
||
| Plant;Flower | Tall Stewartia / Flower / Stewartia monadelpha / Theaceae Stewartia |  |
||
| Plant;Leaf | Nagi, Asian bayberry / Leaf / Nageia nagi / Podocarpaceae Nageia |  |
||
| Plant;Fruit | Nagi, Asian bayberry / Fruit (small) / Nageia nagi / Podocarpaceae Nageia |  |
||
| Plant;Flower | Hydrangea / Flower / Hydrangea macrophylla / Hydrangeaceae Hydrangea |  |
||
| Plant;Leaf | Carolina horse nettle / Leaf / Solanum carolinense / Solanaceae Solanum |  |
||
| Plant;Flower | Carolina horse nettle / Flower / Solanum carolinense / Solanaceae Solanum |  |
||
| Plant;Leaf | Curly dock / Leaf / Rumex crispus / Polygonaceae Rumex |  |
||
| Plant;Flower;Fruit | Curly dock / Flower, Fruit / Rumex crispus / Polygonaceae Rumex |  |
||
| Plant;Leaf | Plume poppy / Leaf / Macleaya cordata / Papaveraceae Macleaya |  |
||
| Plant:Shoot | Sharp dock / Shoot (Leaf, Stem, Flower, Fruit) / Rumex conglomeratus / Polygonaceae Rumex |  |
||
| Plant;Leaf | Japanese false bindweed / Leaf (a bit damaged by feeding) / Calystegia hederacea / Convolvulaceae Calystegia |  |
||
| Plant;Flower | Japanese false bindweed / Flower / Calystegia hederacea / Convolvulaceae Calystegia |  |
||
| Plant;Flower | Bushkiller / Flower / Cayratia japonica / Vitaceae Cayratia |  |
||
| Plant;Leaf | Kudzu / Leaf / Pueraria montana var. lobata / Fabaceae Faboideae Phaseoleae Glycininae Pueraria |  |
||
| Herbal medicine;Plant | Jidabokuippo (Extract Granules) |  |
||
| Plant | Carpet grass / Leaf, Stem, Root (unwashed) / Phyla canescens / Verbenaceae Phyla |  |
||
| Plant;Flower | Carpet grass / Flower / Phyla canescens / Verbenaceae Phyla |  |
||
| Plant:Shoot | Brazilian vervain / Flower, Stem / Verbena brasiliensis / Verbenaceae Verbena |  |
||
| Plant;Flower;Fruit | Bee blossom, Whete gaura / Flower, Flower bud, Fruit / Gaura lindheimeri / Onagraceae Onagroideae Onagreae Gaura |  |
||
| Plant;Leaf | Bee blossom, Whete gaura / Leaf / Gaura lindheimeri / Onagraceae Onagroideae Onagreae Gaura |  |
||
| Plant;Flower | Black-eyed-Susan / Flower / Rudbeckia hirta / Asteraceae Asteroideae Rudbeckia |  |
||
| Plant;Leaf | Black-eyed-Susan / Leaf / Rudbeckia hirta / Asteraceae Asteroideae Rudbeckia |  |
||
| Plant;Flower | Cogongrass, Japanese blood grass / Ear / Imperata cylindrica / Poaceae Imperata |  |
||
| Plant;Flower | Lance-leaved tickseed / Flower / Coreopsis lanceolata / Asteraceae Asteroideae Coreopsis |  |
||
| Plant;Leaf | Lance-leaved tickseed / Leaf / Coreopsis lanceolata / Asteraceae Asteroideae Coreopsis |  |
||
| Plant;Flower | Oleander / Flower, Flower bud / Nerium oleander L. var. indicum / Apocynaceae Apocynoideae Nerium |  |
||
| Plant;Flower | Southern magnolia / Flower / Magnolia grandiflora Linn. / Magnoliaceae Magnolia |  |
||
| Food | Cheese, Processed |  |
||
| Food | Cheese, Gorgonzola |  |
||
| Food | Cheese, Parmigiano reggiano |  |
||
| Food | Cheese, Mimolette |  |
||
| Food | Cheese, White Cheddar |  |
||
| Food | Cheese, Gouda |  |
||
| Food | Cheese, Camembert |  |
||
| Food | Cheese, Mozzarella |  |
||
| Food | Cheese, Brie cheese |  |
||
| Food | Cheese, Blue cheese |  |
||
| Herbal medicine;Plant | Seihaito (Extract Granules) |  |
||
| Plant:Shoot | Ladie's tresses / Shoot, Flower / Spiranthes sinensis var. amoena / Orchidaceae Spiranthes |  |
||
| Plant:Shoot | Yellow bartsia / Shoot, Flower / Bellardia viscosa / Orobanchaceae Bellardia |  |
||
| Plant;Leaf | Persimmon / Leaf / Diospyros kaki Thunb. / Ebenaceae Diospyros |  |
||
| Plant:Shoot | Hop / Leaf (young), Stem / Humulus lupulus / Cannabaceae Humulus |  |
||
| Plant;Flower;Fruit | Plume poppy / Flower, Flower bud, Fruit (immature) / Macleaya cordata / Papaveraceae Macleaya |  |
||
| Plant;Fruit | Tall Stewartia / Fruit (immature) / Stewartia monadelpha / Theaceae Stewartia |  |
||
| Plant:Shoot | Annual fleabane / Leaf, Stem, Flower / Erigeron annuus / Asteraceae Asteroideae Astereae Erigeron |  |
||
| Plant;Flower | Silk tree / Flower / Albizia julibrissin / Fabaceae Mimosoideae Albizia |  |
||
| Plant;Leaf | Silk tree / Leaf / Albizia julibrissin / Fabaceae Mimosoideae Albizia |  |
||
| Plant;Leaf | Chinese tallow tree / Leaf / Triadica sebifera / Euphorbiaceae Euphorbioideae Triadica |  |
||
| Herbal medicine;Plant | Chikujountanto (Extract Granules) |  |
||
| Herbal medicine;Plant | Jiinshihoto (Extract Granules) |  |
||
| Herbal medicine;Plant | Jiinkokato (Extract Granules) |  |
||
| Herbal medicine;Plant | Gokoto (Extract Granules) |  |
||
| Herbal medicine;Plant | Saibokuto (Extract Granules) |  |
||
| Herbal medicine;Plant | Daibofuto (Extract Granules) |  |
||
| Herbal medicine;Plant | Ogikenchuto (Extract Granules) |  |
||
| Herbal medicine;Plant | Shokenchuto (Extract Granules) |  |
||
| Herbal medicine;Plant | Daikenchuto (Extract Granules) |  |
||
| Herbal medicine;Plant | Shomakakkonto (Extract Granules) |  |
||
| Herbal medicine;Plant | Tokito (Extract Granules) |  |
||
| Herbal medicine;Plant | Sansoninto (Extract Granules) |  |
||
| Herbal medicine;Plant | Shin'iseihaito (Extract Granules) |  |
||
| Herbal medicine;Plant | Tsudosan (Extract Granules) |  |
||
| Plant;Leaf | Chinese lantern plant / Leaf / Physalis alkekengi / Solanaceae Physalis |  |
||
| Plant | Chinese lantern plant / Calyx / Physalis alkekengi / Solanaceae Physalis |  |
||
| Plant;Fruit | Chinese lantern plant / Fruit / Physalis alkekengi / Solanaceae Physalis |  |
||
| Plant;Fruit | Nagi, Asian bayberry / Fruit (immature) / Nageia nagi / Podocarpaceae Nageia |  |
||
| Plant;Fruit | Tall Stewartia / Fruit / Stewartia monadelpha / Theaceae Stewartia |  |
||
| Plant;Flower | Marvel of Peru / Flower / Mirabilis jalapa / Nyctaginaceae Mirabilis |  |
||
| Plant;Leaf | Marvel of Peru / Leaf / Mirabilis jalapa / Nyctaginaceae Mirabilis |  |
||
| Plant;Fruit | Marvel of Peru / Fruit / Mirabilis jalapa / Nyctaginaceae Mirabilis |  |
||
| Plant:Shoot | Skunkvine / Flower, Flower bud, Leaf, Stem / Paederia scandens / Rubiaceae Rubioideae Paederieae Paederia |  |
||
| Plant;Fruit | Tall Stewartia / Fruit / Stewartia monadelpha / Theaceae Stewartia |  |
||
| Plant:Shoot | Sweet autumn clematis / Flower, Flower bud, Stem / Clematis terniflora / Ranunculaceae Clematis |  |
||
| Plant;Leaf | Sweet autumn clematis / Leaf / Clematis terniflora / Ranunculaceae Clematis |  |
||
| Standard Chemical | Standard Momilactone A |  |
||
| Standard Chemical | Standard Momilactone B |  |
||
| Plant;Flower | Tomato Micro-Tom (NBRP TOMJPF00001) / Flower / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Flower | Tomato Moneymaker (NBRP TOMJPF00002) / Flower / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Flower | Tomato Aichi First (NBRP TOMJPF00003) / Flower / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Flower | Tomato Ailsa Craig (NBRP TOMJPF00004) / Flower / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Flower | Tomato M82 (NBRP TOMJPF00005) / Flower / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Flower | Tomato Rutgers (NBRP TOMJPF00006) / Flower / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Flower | Tomato Ponderosa (NBRP TOMJPF00007) / Flower / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Flower | Tomato Ueleie106WP (NBRP TOMJPF00015) / Flower / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Flower | Tomato Heinz1706B (NBRP TOMJPF00016) / Flower / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Flower | Tomato S. pennelii (NBRPTOMJPF00008) / Flower / Solanum pennellii / Solanaceae Solanum |  |
||
| Plant;Flower | Tomato Cerasiforme (NBRP TOMJPF00009) / Flower / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Flower | Tomato S. peruvianum 1 (NBRP TOMJPF00010-1) / Flower / Solanum peruvianum / Solanaceae Solanum | 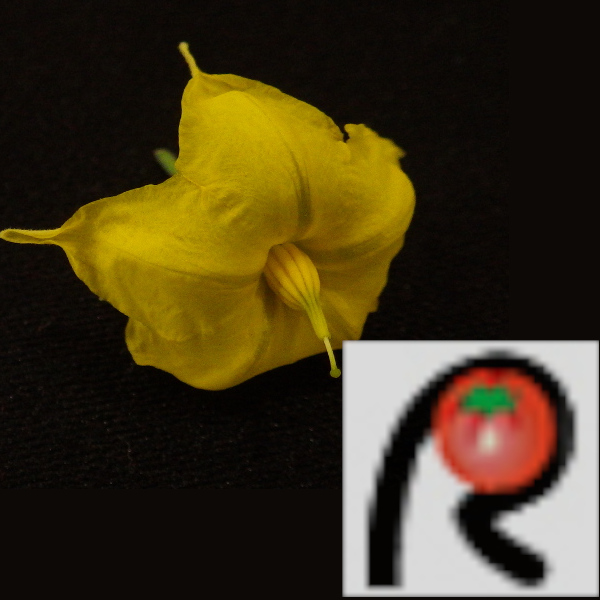 |
||
| Plant;Flower | Tomato S. peruvianum 2 (NBRP TOMJPF00010-2) / Flower / Solanum peruvianum / Solanaceae Solanum |  |
||
| Plant;Flower | Tomato S. peruvianum 3 (NBRP TOMJPF00010-3) / Flower / Solanum peruvianum / Solanaceae Solanum |  |
||
| Plant;Flower | Tomato S. pimpinellifolium 1 (NBRP TOMJPF00012-1) / Flower / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Flower | Tomato S. pimpinellifolium 2 (NBRP TOMJPF00012-2) / Flower / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Flower | Tomato S. pimpinellifolium 3 (NBRP TOMJPF00012-3) / Flower / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Flower | Tomato S. pimpinellifolium 4 (NBRP TOMJPF00012-4) / Flower / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato Micro-Tom (NBRP TOMJPF00001) / Fruit (immature, green) / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato Moneymaker (NBRP TOMJPF00002) / Fruit (immature, green) / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato Aichi First (NBRP TOMJPF00003) / Fruit (immature, green) / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato Ailsa Craig (NBRP TOMJPF00004) / Fruit (immature, green) / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato M82 (NBRP TOMJPF00005) / Fruit (immature, green) / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato Rutgers (NBRP TOMJPF00006) / Fruit (immature, green) / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato Ponderosa (NBRP TOMJPF00007) / Fruit (immature, green) / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato Ueleie106WP (NBRP TOMJPF00015) / Fruit (immature, green) / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato Heinz1706B (NBRP TOMJPF00016) / Fruit (immature, green) / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato S. pennelii (NBRPTOMJPF00008) / Fruit (immature, green) / Solanum pennellii / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato Cerasiforme (NBRP TOMJPF00009) / Fruit (immature, green) / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato S. peruvianum 1 (NBRP TOMJPF00010-1) / Fruit (immature, green) / Solanum peruvianum / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato S. pimpinellifolium 1 (NBRP TOMJPF00012-1) / Fruit (immature, green) / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato S. pimpinellifolium 2 (NBRP TOMJPF00012-2) / Fruit (immature, green) / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato S. pimpinellifolium 3 (NBRP TOMJPF00012-3) / Fruit (immature, green) / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato S. pimpinellifolium 4 (NBRP TOMJPF00012-4) / Fruit (immature, green) / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato Micro-Tom (NBRP TOMJPF00001) / Fruit (mature, red) / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato Moneymaker (NBRP TOMJPF00002) / Fruit (mature, red) / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato Aichi First (NBRP TOMJPF00003) / Fruit (mature, red) / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato Ailsa Craig (NBRP TOMJPF00004) / Fruit (mature, red) / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato M82 (NBRP TOMJPF00005) / Fruit (mature, red) / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato Rutgers (NBRP TOMJPF00006) / Fruit (mature, red) / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato Ponderosa (NBRP TOMJPF00007) / Fruit (mature, red) / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato Ueleie106WP (NBRP TOMJPF00015) / Fruit (mature, red) / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato Heinz1706B (NBRP TOMJPF00016) / Fruit (mature, red) / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato S. pennelii (NBRPTOMJPF00008) / Fruit (mature, red) / Solanum pennellii / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato Cerasiforme (NBRP TOMJPF00009) / Fruit (mature, red) / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato S. peruvianum 1 (NBRP TOMJPF00010-1) / Fruit (mature, red) / Solanum peruvianum / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato S. peruvianum 3 (NBRP TOMJPF00010-3) / Fruit (mature, red) / Solanum peruvianum / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato S. pimpinellifolium 1 (NBRP TOMJPF00012-1) / Fruit (mature, red) / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato S. pimpinellifolium 2 (NBRP TOMJPF00012-2) / Fruit (mature, red) / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato S. pimpinellifolium 3 (NBRP TOMJPF00012-3) / Fruit (mature, red) / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Fruit | Tomato S. pimpinellifolium 4 (NBRP TOMJPF00012-4) / Fruit (mature, red) / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Seed | Tomato Micro-Tom (NBRP TOMJPF00001) / Seed / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Seed | Tomato Moneymaker (NBRP TOMJPF00002) / Seed / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Seed | Tomato Aichi First (NBRP TOMJPF00003) / Seed / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Seed | Tomato Ailsa Craig (NBRP TOMJPF00004) / Seed / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Seed | Tomato M82 (NBRP TOMJPF00005) / Seed / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Seed | Tomato Rutgers (NBRP TOMJPF00006) / Seed / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Seed | Tomato Ponderosa (NBRP TOMJPF00007) / Seed / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Seed | Tomato Ueleie106WP (NBRP TOMJPF00015) / Seed / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Seed | Tomato Heinz1706B (NBRP TOMJPF00016) / Seed / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Seed | Tomato S. pennelii (NBRPTOMJPF00008) / Seed / Solanum pennellii / Solanaceae Solanum |  |
||
| Plant;Seed | Tomato Cerasiforme (NBRP TOMJPF00009) / Seed / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Seed | Tomato S. peruvianum 1 (NBRP TOMJPF00010-1) / Seed / Solanum peruvianum / Solanaceae Solanum |  |
||
| Plant;Seed | Tomato S. peruvianum 2 (NBRP TOMJPF00010-2) / Seed / Solanum peruvianum / Solanaceae Solanum |  |
||
| Plant;Seed | Tomato S. pimpinellifolium 1 (NBRP TOMJPF00012-1) / Seed / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Seed | Tomato S. pimpinellifolium 2 (NBRP TOMJPF00012-2) / Seed / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Seed | Tomato S. pimpinellifolium 3 (NBRP TOMJPF00012-3) / Seed / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Seed | Tomato S. pimpinellifolium 4 (NBRP TOMJPF00012-4) / Seed / Solanum pimpinellifolium / Solanaceae Solanum |  |
||
| Plant;Fruit;Food | Oldham lueberry / Fruit juice / Vaccinium oldhamii / Ericaceae Vaccinoideae Vaccinium |  |
||
| Plant;Flower | A kind of daisy / Flower extract with hot water / Chrysanthemum sp. / Asteraceae Chrysanthemum |  |
||
| Plant;Fruit;Food | Bitter melon / Fruit (lyophilized powder) / Momordica charantia / Cucurbitaceae Momordica |  |
||
| Food;Plant | Vegetable juice 1 |  |
||
| Food;Plant | Vegetable juice 2 |  |
||
| Plant;Flower | Red spider lily / Flower / Lycoris radiata / Amarylidaceae Amarylidoideae Lycorideae Lycoris |  |
||
| Plant;Shoot | Red spider lily / Flower stalk / Lycoris radiata / Amarylidaceae Amarylidoideae Lycorideae Lycoris |  |
||
| Plant;Root | Red spider lily / Scaly bulb / Lycoris radiata / Amarylidaceae Amarylidoideae Lycorideae Lycoris |  |
||
| Plant;Flower | Tall goldenrod / Flower / Solidago canadensis var. scabra / Asteraceae Solidago |  |
||
| Plant;Leaf | Tall goldenrod / Leaf / Solidago canadensis var. scabra / Asteraceae Solidago |  |
||
| Plant;Root | Tall goldenrod / Root (unwashed) / Solidago canadensis var. scabra / Asteraceae Solidago |  |
||
| Plant;Root | Tall goldenrod / Root (washed) / Solidago canadensis var. scabra / Asteraceae Solidago |  |
||
| Food | Nutritional supplement drink 1 |  |
||
| Food | Nutritional supplement drink 2 |  |
||
| Food | Nutritional supplement drink 3 |  |
||
| Plant;Flower | A kind of Lindera / Female flower (old) / Lindera umbellata / Lauraceae Lindera |  |
||
| Plant;Flower | A kind of Lindera / Male flower (old) / Lindera umbellata / Lauraceae Lindera |  |
||
| Plant;Leaf | A kind of Lindera / Leaf / Lindera umbellata / Lauraceae Lindera |  |
||
| Plant;Shoot | A kind of Lindera / Bark (greenish) / Lindera umbellata / Lauraceae Lindera |  |
||
| Plant;Shoot | A kind of Lindera / Bark (blackish) / Lindera umbellata / Lauraceae Lindera |  |
||
| Product | Distilled water when making the essential oil of Lindera umbellata / Lindera umbellata / Lauraceae Lindera |  |
||
| Product | Essential oil of Lindera umbellata / Lindera umbellata / Lauraceae Lindera |  |
||
| Herbal medicine | Kampo stomach medicine 1 |  |
||
| Herbal medicine | Kampo stomach medicine 2 |  |
||
| Herbal medicine | Kampo folk medicine |  |
||
| Herbal medicine;Food | Kampo troche |  |
||
| Food;Plant | Barley drink 1 |  |
||
| Food;Plant | Barley drink 2 |  |
||
| Food;Plant | Barley drink 3 |  |
||
| Plant;Leaf | Dalmatian chrysanthemum (NBRP YSY01) / Leaf / Tanacetum cinerariifolium / Asteraceae Asteroideae Tanacetum |  |
||
| Plant;Flower | Dalmatian chrysanthemum (NBRP YSY01) / Flower / Tanacetum cinerariifolium / Asteraceae Asteroideae Tanacetum |  |
||
| Plant;Leaf | Iwainchin (a kind of chrysanthemum, NBRP AMJ06) / Leaf / Chrysanthemum rupestre Matsum / Asteraceae Asteroideae Chrysanthemum |  |
||
| Plant;Flower | Iwainchin (a kind of chrysanthemum, NBRP AMJ06) / Flower / Chrysanthemum rupestre Matsum / Asteraceae Asteroideae Chrysanthemum |  |
||
| Plant;Leaf | Pireo-giku (a kind of chrysanthemum, NBRP YCF01) / Leaf / Chrysanthemum weyrichii / Asteraceae Asteroideae Chrysanthemum |  |
||
| Plant;Flower | Pireo-giku (a kind of chrysanthemum, NBRP YCF01) / Flower / Chrysanthemum weyrichii / Asteraceae Asteroideae Chrysanthemum |  |
||
| Product | Mosquito repellent 1 |  |
||
| Product | Mosquito repellent 2 |  |
||
| Product | Mosquito repellent, ash 1 |  |
||
| Product | Mosquito repellent, ash 2 |  |
||
| Plant;Seed | Oat / Seed / Avena sativa / Poaceae Avena |  |
||
| Marine algae | Umiuchiwa seaweed, Sea-fan / Padina arborescens Holmes / Dictyotaceae Padina |  |
||
| Marine algae | Nebarimo seaweed / Leathesia marina (Lyngbye) Decaisne / Chordariaceae Leathesia |  |
||
| Marine algae | Iroro seaweed / Ishige foliacea Okamura / Ishigeaceae Ishige |  |
||
| Marine algae | Ishige seaweed / Ishige okamurae Yendo / Ishigeaceae Ishige |  |
||
| Marine algae | Wakame seaweed (young) / Frond (young) / Undaria pinnatifida (Harvey) Suringar / Alariaceae Undaria |  |
||
| Marine algae | Wakame seaweed / Fertile frond (young) / Undaria pinnatifida (Harvey) Suringar / Alariaceae Undaria |  |
||
| Marine algae | Wakame seaweed / Fertile frond / Undaria pinnatifida (Harvey) Suringar / Alariaceae Undaria |  |
||
| Marine algae | Arame seaweed / Frond / Eisenia bicyclis (Kjellman) Setchell / Lessoniaceae Eisenia |  |
||
| Marine algae | Arame seaweed / Axis / Eisenia bicyclis (Kjellman) Setchell / Lessoniaceae Eisenia |  |
||
| Marine algae | Akamoku seaweed / Sargassum horneri (Turner) C. Agardh / Sargassaceae Sargassum |  |
||
| Marine algae | Akamoku seaweed / Alga body with many female reproductive organs / Sargassum horneri (Turner) C. Agardh / Sargassaceae Sargassum |  |
||
| Marine algae | Akamoku seaweed / Alga body with many air bladders / Sargassum horneri (Turner) C. Agardh / Sargassaceae Sargassum |  |
||
| Herbal medicine;Plant | Unkeito (Extract Granules) |  |
||
| Herbal medicine;Plant | Goshajinkigan (Extract Granules) |  |
||
| Herbal medicine;Plant | Ninjin'yoeito (Extract Granules) |  |
||
| Herbal medicine;Plant | Shosaikotokakikyosekko (Extract Granules) |  |
||
| Marine algae | Hijiki seaweed / Sargassum fusiforme (Harvey) Setchell / Sargassaceae Sargassum |  |
||
| Marine algae | Hijiki seaweed / Alga body (young) / Sargassum fusiforme (Harvey) Setchell / Sargassaceae Sargassum |  |
||
| Marine algae | Oobamoku seaweed / Sargassum ringgoldianum Harvey ssp. ringgoldianum / Sargassaceae Sargassum |  |
||
| Marine algae | Oobamoku seaweed / Axis / Sargassum ringgoldianum Harvey ssp. ringgoldianum / Sargassaceae Sargassum |  |
||
| Marine algae | Hiranejimoku seaweed / Sargassum okamurae Yoshida et T. Konno / Sargassaceae Sargassum |  |
||
| Marine algae | Narasamo seaweed / Sargassum nigrifolium Yendo / Sargassaceae Sargassum |  |
||
| Marine algae | Umitoranoo seaweed / Sargassum thunbergii (Mertens ex Roth) Kuntze / Sargassaceae Sargassum |  |
||
| Marine algae | Heritorikaninote seaweed / Corallina crassissima (Yendo) Hind et Saunders / Corallinaceae Corallina |  |
||
| Marine algae | Pirihiba seaweed 1 / Corallina pilulifera Postels et Ruprecht / Corallinaceae Corallina |  |
||
| Marine algae | Pirihiba seaweed 2 / Corallina pilulifera Postels et Ruprecht / Corallinaceae Corallina |  |
||
| Marine algae | Pirihiba seaweed 3 / Corallina pilulifera Postels et Ruprecht / Corallinaceae Corallina |  |
||
| Marine algae | Sabimodoki seaweed / Corallina melobesioides (Segawa) Martone, Lindstrom, Miller et Gabrielson / Corallinaceae Corallina |  |
||
| Food;Fermented;Plant | Pickled scallions / Allium chinense |  |
||
| Food;Fermented;Plant | Black garlic / Allium sativum |  |
||
| Herbal medicine;Plant | Rikkosan (Extract Granules) |  |
||
| Medium | Medium (for Duckweed, Lemna aequinoctialis) |  |
||
| Plant | Duckweed / Aceptic culture, 10 days / Lemna aequinoctialis T.Beppu & Murata, 1985 subsp. aoukikusa / Araceae Lemnoideae Lemna |  |
||
| Herbal medicine;Plant | Seishinrenshiin (Extract Granules) |  |
||
| Herbal medicine;Microbes;Fungi | Chinese caterpillar fungus product 1 / Ophiocordyceps sinensis (Berkeley) Saccardo / Ophiocordycipitaceae Ophiocordyceps |  |
||
| Herbal medicine;Microbes;Fungi | Chinese caterpillar fungus product 2 / Ophiocordyceps sinensis (Berkeley) Saccardo / Ophiocordycipitaceae Ophiocordyceps |  |
||
| Herbal medicine;Microbes;Fungi | Chinese caterpillar fungus product 3 / Ophiocordyceps sinensis (Berkeley) Saccardo / Ophiocordycipitaceae Ophiocordyceps |  |
||
| Herbal medicine;Microbes;Fungi | Chinese caterpillar fungus product 4 / Ophiocordyceps sinensis (Berkeley) Saccardo / Ophiocordycipitaceae Ophiocordyceps |  |
||
| Standard Chemical | Standard Adenine |  |
||
| Standard Chemical | Standard mix. / Cytosine; Thymine; Uracil |  |
||
| Standard Chemical | Standard Guanine |  |
||
| Standard Chemical | Standard Adenosine |  |
||
| Standard Chemical | Standard mix. / Cytidine; Guanosine; Thymidine; Uridine |  |
||
| Standard Chemical | Standard 2'-Deoxyadenosine |  |
||
| Standard Chemical | Standard Cordycepin; 3'-Deoxyadenosine |  |
||
| Standard Chemical | Standard mix. / dATP (Deoxyadenosine triphosphate); dCTP (Deoxycytidine triphosphate); dGTP (Deoxyguanosine triphosphate); dTTP (Deoxythymidine triphosphate) |  |
||
| Standard Chemical | Standard mix. / ATP (Adenosine triphosphate); CTP (Cytidine triphosphate); GTP (Guanosine triphosphate); UTP (Uridine triphosphate) |  |
||
| Herbal medicine;Plant | Choreitogoshimotsuto (Extract Granules) |  |
||
| Marine algae | Maoukaninote seaweed / Amphiroa ephedraea (Lamarck) Decaisne / Corallinaceae Amphiroa |  |
||
| Marine algae | Musetsusangomo seaweed / Corallinaceae |  |
||
| Marine algae | Makusa seaweed 1 / Gelidium elegans Kuezing / Gelidiaceae Gelidium |  |
||
| Marine algae | Makusa seaweed 2 / Gelidium elegans Kuezing / Gelidiaceae Gelidium |  |
||
| Marine algae | Onikusa seaweed / Gelidium japonicum (Haravey) Okamura / Gelidiaceae Gelidium |  |
||
| Marine algae | Hanafunori seaweed / Gloiopeltis complanata (Harvey) Yamada / Endocladiaceae Gloiopeltis |  |
||
| Marine algae | Fukurofunori seaweed / Gloiopeltis furcata (Postels et Ruprecht) J. Agardh / Endocladiaceae Gloiopeltis |  |
||
| Marine algae | Suginori seaweed / Chondracanthus tenellus (Harvey) Hommersand / Gigartinaceae Chondracanthus |  |
||
| Marine algae | Kainori seaweed / Chondracanthus intermedius (Suringar) Hommersand / Gigartinaceae Chondracanthus |  |
||
| Marine algae | Ibotsunomata seaweed / Alga body (young) without cysts / Chondrus verrucosus Mikami / Gigartinaceae Chondrus |  |
||
| Herbal medicine;Plant | San'oshashinto (Extract Granules) |  |
||
| Standard Chemical | Standard (+)-Limonene |  |
||
| Standard Chemical | Standard (-)-Limonene |  |
||
| Standard Chemical | Standard (-)-Menthol |  |
||
| Standard Chemical | Standard Vincristine |  |
||
| Standard Chemical | Standard Capsaicin |  |
||
| Herbal medicine;Plant | Saireito (Extract Granules) |  |
||
| Marine algae | Ibotsunomata seaweed / Alga body (greenish) / Chondrus verrucosus Mikami / Gigartinaceae Chondrus |  |
||
| Marine algae | Ibotsunomata seaweed / Alga body with cysts / Chondrus verrucosus Mikami / Gigartinaceae Chondrus |  |
||
| Marine algae | Kagiibaranori seaweed / Hypnea japonica Tanaka / Hypneaceae Hypnea |  |
||
| Marine algae | Hanesaimi seaweed / Ahnfeltiopsis yamadae (Segawa) Masuda / Phyllophoraceae Ahnfeltiopsis |  |
||
| Marine algae | Harigane seaweed / Ahnfeltiopsis paradoxa (Suringar) Masuda / Phyllophoraceae Ahnfeltiopsis |  |
||
| Marine algae | Hosobanaminohana seaweed / Portieria hornemannii (Lyngbye) Silva / Rhizophyllidaceae Portieria |  |
||
| Marine algae | Kintoki seaweed / Grateloupia angusta (Okamura) Kawaguchi et Wang / Halymeniaceae Grateloupia |  |
||
| Marine algae | Matsunori seaweed / Polyopes affinis (Harvey) Kawaguchi et Wang / Halymeniaceae Polyopes |  |
||
| Marine algae | Hizirimen seaweed / Grateloupia sparsa (Okamura) Chiang / Halymeniaceae Grateloupia |  |
||
| Marine algae | Tanbanori seaweed / Pachymeniopsis elliptica (Holmes) Yamada / Halymeniaceae Pachymeniopsis |  |
||
| Herbal medicine;Plant | Ireito (Extract Granules) |  |
||
| Food;Plant;Fruit | Red pepper / Fruit (dried) / Capsicum annuum / Solanaceae Capsicum |  |
||
| Food;Plant | Habanero chilli (dried powder) / Capsicum chinense / Solanaceae Capsicum |  |
||
| Food;Plant | Japanese pepper (dried powder) / Zanthoxylum piperitum / Rutaceae Zanthoxylum |  |
||
| Food;Plant | Sichuan pepper (dried powder) / Zanthoxylum bungeanum / Rutaceae Zanthoxylum |  |
||
| Product;Plant | Essential oil Ylang ylang / Flower (Essential oil) / Cananga odorata / Annonaceae Cananga |  |
||
| Product;Plant | Essential oil Clary sage / Flower, Leaf (Essential oil) / Salvia sclarea / Lamiaceae Salvia |  |
||
| Product;Plant | Essential oil Grapefruit / Peel (Essential oil) / Citrus x paradisi / Rutaceae Citrus |  |
||
| Product;Plant | Essential oil Juniperberry / Fruit (Essential oil) / Juniperus communis / Cupressaceae Juniperus |  |
||
| Product;Plant | Essential oil Sweet orange / Peel (Essential oil) / Citrus sinensis / Rutaceae Citrus |  |
||
| Product;Plant | Essential oil Sweet marjoram / Leaf (Essential oil) / Origanum majorana / Lamiaceae Origanum |  |
||
| Product;Plant | Essential oil Geranium / Flower, Leaf (Essential oil) / Pelargonium graveolens / Geraniaceae Pelargonium |  |
||
| Product;Plant | Essential oil Tea tree / Leaf (Essential oil) / Melaleuca alternifolia / Myrtaceae Melaleuca |  |
||
| Product;Plant | Essential oil Frankincense / Resin (Essential oil) / Boswellia carterii / Burseraceae Boswellia |  |
||
| Product;Plant | Essential oil Peppermint / Leaf (Essential oil) / Mentha x piperita / Lamiaceae Mentha |  |
||
| Product;Plant | Essential oil Bergamot / Peel (Essential oil) / Citrus x bergamia / Rutaceae Citrus |  |
||
| Product;Plant | Essential oil Eucalyptus / Leaf (Essential oil) / Eucalyptus globulus / Myrtaceae Eucalyptus |  |
||
| Product;Plant | Essential oil Lavender / Flower, Leaf (Essential oil) / Lavandula angustifolia / Lamiaceae Lavandula |  |
||
| Product;Plant | Essential oil Lemon / Peel (Essential oil) / Citrus limon / Rutaceae Citrus |  |
||
| Product;Plant | Essential oil Lemongrass / Leaf (Essential oil) / Cymbopogon flexuosus / Poaceae Cymbopogon |  |
||
| Product;Plant | Essential oil Rosemary cineole / Leaf (Essential oil) / Rosmarinus officinalis L. / Lamiaceae Rosmarinus |  |
||
| Product;Plant | Essential oil Roman Chamomile / Flower (Essential oil) / Chamaemelum nobile / Asteraceae Asteroideae Anthemideae Chamaemelum |  |
||
| Standard Chemical | Standard beta-Citronellol |  |
||
| Standard Chemical | Standard Geraniol |  |
||
| Standard Chemical | Standard Linalool |  |
||
| Standard Chemical | Standard Citral |  |
||
| Standard Chemical | Standard Nerol |  |
||
| Standard Chemical | Standard alpha-Terpineol |  |
||
| Food;Plant;Seed | Purple-black rice (Asamurasaki) / Grain, raw (brown rice) / Oryza sativa / Poaceae Oryzoideae Oryza |  |
||
| Food;Plant | Rice oil (Solvent extract) / Oryza sativa / Poaceae Oryzoideae Oryza |  |
||
| Food;Plant | Rice oil (Compressed) / Oryza sativa / Poaceae Oryzoideae Oryza |  |
||
| Food;Plant | Cake flour / Triticum aestivum / Poaceae Triticum |  |
||
| Food | Tempura flour |  |
||
| Medium | AG medium (for Azospirillum sp., unused) |  |
||
| Medium | AG medium supernatant (after culturing Azospirillum sp. B510) / Azospirillum sp. B510 / Rhodospirillaceae Azospirillum |  |
||
| Microbes | Azospirillum sp. B510 / Cell / Azospirillum sp. B510 / Rhodospirillaceae Azospirillum |  |
||
| Medium | MSC medium (for Methylobacterium indicum, unused) |  |
||
| Medium | MSC medium supernatant (after culturing Methylobacterium indicum SE2.11) / Methylobacterium indicum SE2.11 / Methylobacteriaceae Methylobacterium |  |
||
| Microbes | Methylobacterium indicum SE2.11 / Cell / Methylobacterium indicum SE2.11 / Methylobacteriaceae Methylobacterium |  |
||
| Herbal medicine;Plant | Bukuryoingohangekobokuto (Extract Granules) |  |
||
| Marine algae | Hitotsumatsu seaweed / Grateloupia chiangii Kawaguchi et Wang / Halymeniaceae Grateloupia |  |
||
| Marine algae | Benisunago seaweed / Schizymenia dubyi (Chauvin ex Duby) J. Agardh / Schizymeniaceae Schizymenia |  |
||
| Marine algae | Iwanokawa seaweed / Peyssonnelia sp. / Peyssonneliaceae Peyssonnelia |  |
||
| Marine algae | Ogonori seaweed / Gracilaria vermiculophylla (Ohmi) Papenfuss / Gracilariaceae Gracilaria |  |
||
| Marine algae | Ogonori seaweed / Alga body with cysts / Gracilaria vermiculophylla (Ohmi) Papenfuss / Gracilariaceae Gracilaria |  |
||
| Marine algae | Mizoogonori seaweed / Gracilaria incurvata Okamura / Gracilariaceae Gracilaria |  |
||
| Marine algae | Fushitsunagi seaweed / Lomentaria catenata Harvey / Lomentariaceae Lomentaria |  |
||
| Marine algae | Magiresozo seaweed / Laurencia saitoi Perestenko / Rhodomelaceae Laurencia |  |
||
| Marine algae | Kamogashiranori seaweed / Dermonema pulvinatum (Grunow ex Holmes) Fan / Liagoraceae Dermonema |  |
||
| Marine algae | Benimadara seaweed / Hildenbrandia rubra (Sommerfelt) Meneghini / Hildenbrandiaceae Hildenbrandia |  |
||
| Marine algae | Hitoegusa seaweed / Monostroma nitidum Wittrock / Gomontiaceae Monostroma |  |
||
| Marine algae | Sujiaonori seaweed (including sand) / Ulva prolifera O.F. Müller / Ulvaceae Ulva |  |
||
| Marine algae | Botan-aosa seaweed / Ulva conglobata Kjellman / Ulvaceae Ulva |  |
||
| Marine algae | Anaaosa seaweed / Ulva pertusa Kjellman / Ulvaceae Ulva |  |
||
| Marine algae | Kaigoromo seaweed / Pseudocladophora conchopheria (Sakai) Boedeker et Leliaert / Cladophoraceae Pseudocladophora |  |
||
| Animal | Daidaiisokaimen (a kind of sponge) / Halichondria japonica / Halichondriidae Hymeniacidon |  |
||
| Animal | Siroboya (a kind of sea squirt) / Styela plicata / Styelidae Styelinae Styela |  |
||
| Animal | Vase tunicate (a kind of sea squirt) / Sperm / Ciona intestinalis type A or Ciona robusta / Cionidae Ciona |  |
||
| Animal | Vase tunicate (a kind of sea squirt) / Egg / Ciona intestinalis type A or Ciona robusta / Cionidae Ciona |  |
||
| Food;Alcoholic beverage | Whisky 1 (Japanese) |  |
||
| Food;Alcoholic beverage | Beer 1 (Japanese) |  |
||
| Food;Alcoholic beverage | Beer 2 (Japanese) |  |
||
| Food;Alcoholic beverage | Beer 3 (Japanese) |  |
||
| Food;Alcoholic beverage | Beer 4 (Japanese) |  |
||
| Food;Alcoholic beverage | Beer 5 (Japanese) |  |
||
| Food;Alcoholic beverage | Whisky 2 (Scotch) |  |
||
| Food;Alcoholic beverage | Whisky 3 (Scotch) |  |
||
| Food;Alcoholic beverage | Whisky 4 (American) |  |
||
| Food;Alcoholic beverage | Whisky 5 (Scotch) |  |
||
| Food;Alcoholic beverage | Whisky 6 (Scotch) |  |
||
| Food;Alcoholic beverage | Whisky 7 (Scotch) |  |
||
| Food;Alcoholic beverage | Whisky 8 (Scotch) |  |
||
| Food;Alcoholic beverage | Whisky 9 (Scotch) |  |
||
| Food;Alcoholic beverage | Whisky 10 (Scotch) |  |
||
| Plant;Leaf | Common gromwell, European stoneseed 1 / Leaf / Lithospermum officinale / Boraginaceae Lithospermum |  |
||
| Plant;Leaf | Common gromwell, European stoneseed 2 / Leaf / Lithospermum officinale / Boraginaceae Lithospermum |  |
||
| Plant;Leaf | Red-root gromwell / Leaf / Lithospermum erythrorhizon / Boraginaceae Lithospermum |  |
||
| Food;Plant;Seed | Ginkgo / Seed (Ginkgo nut, raw) / Ginkgo biloba / Ginkgoaceae Ginkgo |  |
||
| Food;Plant;Seed | Maize / Seed / Zea mays subsp. mays / Poaceae Zea |  |
||
| Food;Microbes;Fungi | Ear fungus (dried) / Auricularia sp. (Auricularia auricula-judae) / Auricularaceae Auricularia |  |
||
| Plant;Leaf | Tomato Micro-Tom, Mock control 2 days / Leaf / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Plant;Leaf | Tomato Micro-Tom, Methyl jasmonate (MeJA) treatment 2 days / Leaf / Solanum lycopersicum / Solanaceae Solanum |  |
||
| Standard Chemical | Standard Tomatidine |  |
||
| Standard Chemical | Standard beta-Sitosterol |  |
||
| Standard Chemical | Standard Campesterol |  |
||
| Plant;Shoot;Food | Angelica tenuisecta var. furcijuga (powder product) / Leaf, Stem / Angelica tenuisecta (Makino) Makino var. furcijuga (Kitag.) H.Ohba / Apiaceae Angelica |  |
||
| Standard Chemical | Standard Bergapten |  |
||
| Standard Chemical | Standard 4-Hydroxycoumarin |  |
||
| Standard Chemical | Standard Chlorogenic acid |  |
||
| Standard Chemical | Standard Kaempferol |  |
||
| Standard Chemical | Standard Luteolin |  |
||
| Standard Chemical | Standard Apigenin |  |
||
| Standard Chemical | Standard Folic acid |  |
||
| Standard Chemical | Standard Rosmarinic acid |  |
||
| Standard Chemical | Standard Naringin |  |
||
| Standard Chemical | Standard Allyl isothiocyanate |  |
||
| Standard Chemical | Standard Methyl isothiocyanate |  |
||
| Standard Chemical | Standard Sulforaphane |  |
||
| Standard Chemical | Standard Phenyl isothiocyanate |  |
||
| Standard Chemical | Standard Benzyl isothiocyanate |  |
||
| Herbal medicine;Plant | Unchingoreisan (Extract Granules) |  |
||
| Herbal medicine;Plant | Ryokyojutsukanto (Extract Granules) |  |
||
| Herbal medicine;Plant | Ryokankyomishingeninto (Extract Granules) |  |
||
| Herbal medicine;Plant | Orento (Extract Granules) |  |
||
| Herbal medicine;Plant | Sammotsuogonto (Extract Granules) |  |
||
| Herbal medicine;Plant | Hainosankyuto (Extract Granules) |  |
||
| Herbal medicine;Plant | Tokikenchuto (Extract Granules) |  |
||
| Herbal medicine;Plant | Senkyuchachosan (Extract Granules) |  |
||
| Herbal medicine;Plant | Keishibukuryogankayokuinin (Extract Granules) |  |
||
| Standard Chemical | Standard 7,8-dihydroxycoumarin (Daphnetin) |  |
||
| Standard Chemical | Standard 4,7-dihydroxycoumarin |  |
||
| Standard Chemical | Standard 6,7-dihydroxycoumarin (Esculetin) |  |
||
| Standard Chemical | Standard 2-Oxoglutarate (2-Ketoglutaric acid) |  |
||
| Standard Chemical | Standard cis-Aconitic acid |  |
||
| Standard Chemical | Standard p-Coumaric acid |  |
||
| Standard Chemical | Standard Flavin mononucleotide (FMN) |  |
||
| Plant;Leaf | Japanese cedar / Leaf / Cryptomeria japonica / Cupressaceae Cryptomeria |  |
||
| Plant;Shoot | Japanese cedar / Heartwood / Cryptomeria japonica / Cupressaceae Cryptomeria |  |
||
| Plant;Shoot | Japanese cedar / Sapwood / Cryptomeria japonica / Cupressaceae Cryptomeria |  |
||
| Environment | Wastewater, untreated (lyophilized) |  |
||
| Environment | Wastewater, yeast treated (lyophilized) |  |
||
| Environment | Sludge of wastewater treatment (lyophilized) |  |
||
| Microbes;Microalgae | Nannochloropsis granulata (NBRC 102713) / Cell (grown in F2N medium) / Nannochloropsis granulata / Monodopsidaceae Nannochloropsis |  |
||
| Microbes;Microalgae | Microchloropsis salina (NBRC 102719, Nannochloropsis) / Cell (grown in F2N medium) / Microchloropsis salina (Nannochloropsis salina) / Monodopsidaceae Microchloropsis (Nannochloropsis) |  |
||
| Microbes;Microalgae | Nannochloropsis oceanica (NBRC 102738) / Cell (grown in F2N medium) / Nannochloropsis oceanica / Monodopsidaceae Nannochloropsis |  |
||
| Microbes;Microalgae | Nannochloropsis gaditana (NIES-2587, Microchloropsis) / Cell (grown in F2N medium) / Nannochloropsis gaditana (Microchloropsis gaditana) / Monodopsidaceae Nannochloropsis (Microchloropsis) |  |
||
| Microbes;Microalgae | Nannochloropsis granulata (NIES-2588) / Cell (grown in F2N medium) / Nannochloropsis granulata / Monodopsidaceae Nannochloropsis |  |
||
| Microbes;Microalgae | Nannochloropsis oceanica (NIES-2146) / Cell (grown in F2N medium) / Nannochloropsis oceanica / Monodopsidaceae Nannochloropsis |  |
||
| Microbes;Microalgae | Nannochloropsis oceanica (NIES-2145) / Cell (grown in F2N medium) / Nannochloropsis oceanica / Monodopsidaceae Nannochloropsis |  |
||
| Medium | F2N medium (for Nannochloropsis sp., unused) |  |
||
| Microbes;Microalgae | Euglena gracilis (Euglena Co., Ltd., eu029) / Cell (grown in CM medium pH3.5) / Euglena gracilis / Euglenaceae Englena |  |
||
| Microbes;Microalgae | Parachlorella kessleri (NIES-2152) / Cell (grown in BG11 medium) / Parachlorella kessleri / Chlorellaceae Parachlorella |  |
||
| Microbes;Microalgae | Chlorella sorokiniana (UTEX1230) / Cell (grown in BG11 medium) / Chlorella sorokiniana / Chlorellaceae Chlorella |  |
||
| Microbes;Microalgae | Scenedesmus obliquus (UTEX393) / Cell (grown in BG11 medium) / Scenedesmus obliquus / Scenedesmaceae Scenedesmus |  |
||
| Microbes | Lipomyces starkeyi (NBRC 10381) / Cell / Lipomyces starkeyi / Lipomycetaceae Lipomyces |  |
||
| Standard Chemical | Standard Quercetin 3-O-beta-D-glucuronide |  |
||
| Standard Chemical | Standard 5-Methyl furfural |  |
||
| Standard Chemical | Standard Morin |  |
||
| Standard Chemical | Standard Glyceollin IV |  |
||
| Standard Chemical | Standard (+/-)-8-Prenylnaringenin |  |
||
| Standard Chemical | Standard Yohimbine |  |
||
| Microbes;Archea | Thermococcus kodakarensis (NBRP, JCM 12380) / Cell (grown in medium No.280, JCM) / Thermococcus kodakarensis / Thermococcaceae Thermococcus |  |
||
| Microbes;Archea | Pyrococcus furiosus (NBRP, JCM 8422) / Cell (grown in medium No.151, JCM) / Pyrococcus furiosus / Thermococcaceae Pyrococcus |  |
||
| Microbes;Archea | Methanocaldococcus indicus (NBRP, JCM 11886) / Cell (grown in medium No.232, JCM) / Methanocaldococcus indicus / Methanocaldococcaceae Methanocaldococcus |  |
||
| Microbes;Archea | Methanocaldococcus jannaschii (NBRP, JCM 10045) / Cell (grown in medium No.232, JCM) / Methanocaldococcus jannaschii / Methanocaldococcaceae Methanocaldococcus |  |
||
| Medium | Medium (JCM No.280, Thermococcus medium) |  |
||
| Medium | Medium (JCM No.151, Pyrococcus medium) |  |
||
| Medium | Medium (JCM No.232, Mthanocaldococcus jannaschii medium) |  |
||
| Microbes;Yeast | Saccharomyces cerevisiae (NBRP BY611) / Cell (grown in YPDA medium) / Saccharomyces cerevisiae / Saccharomycetaceae Saccharomyces |  |
||
| Microbes;Yeast | Saccharomyces cerevisiae (NBRP BY4502) / Cell (grown in YPDA medium) / Saccharomyces cerevisiae / Saccharomycetaceae Saccharomyces |  |
||
| Microbes;Yeast | Saccharomyces cerevisiae (NBRP BY29039) / Cell (grown in YPDA medium) / Saccharomyces cerevisiae / Saccharomycetaceae Saccharomyces |  |
||
| Medium | Medium (YPDA) |  |
||
| Standard Chemical | Standard Orysastrobin |  |
||
| Standard Chemical | Standard Trigonelline |  |
||
| Food;Alcoholic beverage | Wine 1 (White, Japan) |  |
||
| Food;Alcoholic beverage | Wine 2 (White, Japan) |  |
||
| Food;Alcoholic beverage | Wine 3 (White, Japan) |  |
||
| Food;Alcoholic beverage | Wine 4 (White, Japan) |  |
||
| Food;Alcoholic beverage | Wine 5 (White, Europe) |  |
||
| Food;Alcoholic beverage | Japanese Sake 1 (Junmai Ginjo) |  |
||
| Standard Chemical | Standard Apigenin-7-O-beta-glucoside |  |
||
| Standard Chemical | Standard Kaempferol-3-O-beta-glucoside |  |
||
| Standard Chemical | Standard Quercetin-3-O-beta-glucoside |  |
||
| Standard Chemical | Standard Juglone (5-hydroxy-1,4-naphtalenedione; 5-hydroxynaphthoquinone) |  |
||
| Standard Chemical | Standard 6-methoxy-2-benzoxazolinone (MBOA) |  |
||
| Standard Chemical | Standard Glycitein |  |
||
| Plant;Shoot | Rice IR64 / Hull / Oryza sativa L. cv IR64 / Poaceae Oryzoideae Oryza |  |
||
| Herbal medicine;Plant | Mashiningan (Extract Granules) |  |
||
| Herbal medicine;Plant | Maobushisaishinto (Extract Granules) |  |
||
| Herbal medicine;Plant | Keihito (Extract Granules) |  |
||
| Herbal medicine;Plant | Daijokito (Extract Granules) |  |
||
| Herbal medicine;Plant | Keishikashakuyakudaioto (Extract Granules) |  |
||
| Herbal medicine;Plant | Inchinkoto (Extract Granules) |  |
||
| Herbal medicine;Plant | Seishoekkito (Extract Granules) |  |
||
| Herbal medicine;Plant | Kamikihito (Extract Granules) |  |
||
| Herbal medicine;Plant | Kikyoto (Extract Granules) |  |
||
| Plant;Flower | Anemone / Flower / Anemone coronaria / Ranunculaceae Anemone |  |
||
| Plant;Flower | Iceland poppy / Flower / Papaver nudicaule / Papaveraceae Papaver |  |
||
| Plant;Flower | Columbine / Flower / Aquilegia coerulea / Ranunculaceae Aquilegia |  |
||
| Plant;Flower | Daffodil / Flower / Narcissus cyclamineus 'Tete a tete' / Amaryllidaceae Amaryllidoideae Narcissus |  |
||
| Plant;Leaf | Dill / Leaf, Stem / Anethum graveolens / Apiaceae Anethum |  |
||
| Plant;Leaf | Digitalis (Foxglove) / Leaf / Digitalis purpurea L. / Plantaginaceae Digitalis |  |
||
| Plant;Leaf | Anemone / Leaf / Anemone coronaria / Ranunculaceae Anemone |  |
||
| Plant;Shoot | Columbine / Leaf, Petiole / Aquilegia coerulea / Ranunculaceae Aquilegia |  |
||
| Plant;Leaf | Daffodil / Leaf / Narcissus cyclamineus 'Tete a tete' / Amaryllidaceae Amaryllidoideae Narcissus |  |
||
| Plant;Root | Dill / Root (washed) / Anethum graveolens / Apiaceae Anethum |  |
||
| category | Columbine / Root (washed) / Aquilegia coerulea / Ranunculaceae Aquilegia |  |
||
| category | Digitalis (Foxglove) / Root (washed) / Digitalis purpurea L. / Plantaginaceae Digitalis |  |
||
| category | Daffodil / Root (washed) / Narcissus cyclamineus 'Tete a tete' / Amaryllidaceae Amaryllidoideae Narcissus |  |
||
| category | Anemone / Root (washed) / Anemone coronaria / Ranunculaceae Anemone |  |
||
| Plant;Leaf | Artichoke / Leaf / Cynara scolymus / Asteraceae Cynareae Cynara |  |
||
| Plant;Leaf | Iceland poppy / Leaf / Papaver nudicaule / Papaveraceae Papaver |  |
||
| Plant;Flower | Lupin / Flower / Lupinus / Fabaceae Lupinus |  |
||
| Plant;Leaf | Lupin / Leaf / Lupinus / Fabaceae Lupinus |  |
||
| category | Artichoke / Root (washed) / Cynara scolymus / Asteraceae Cynareae Cynara |  |
MassChroViewer tool
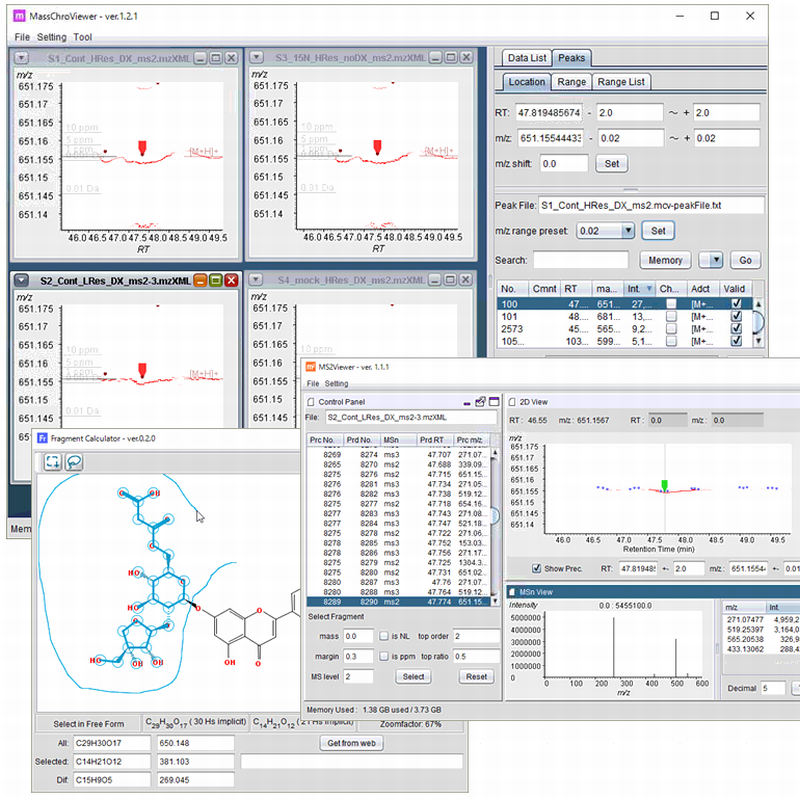
|
The mzXML data and the peak list data available above can be opened by the MassChroViewer software.
Detailed information, such as exact peak position, raw MSn data for each scan can be seen,
and further annotation looking at the chemical structures of candidate compounds can be performed with
the tool. MassChroViewer is available free of charge for academic purposes. |